NetMHCpan, a method for MHC class I binding prediction beyond humans
- PMID: 19002680
- PMCID: PMC3319061
- DOI: 10.1007/s00251-008-0341-z
NetMHCpan, a method for MHC class I binding prediction beyond humans
Abstract
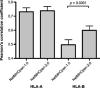
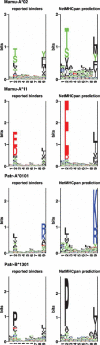
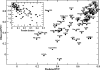
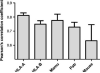
Binding of peptides to major histocompatibility complex (MHC) molecules is the single most selective step in the recognition of pathogens by the cellular immune system. The human MHC genomic region (called HLA) is extremely polymorphic comprising several thousand alleles, each encoding a distinct MHC molecule. The potentially unique specificity of the majority of HLA alleles that have been identified to date remains uncharacterized. Likewise, only a limited number of chimpanzee and rhesus macaque MHC class I molecules have been characterized experimentally. Here, we present NetMHCpan-2.0, a method that generates quantitative predictions of the affinity of any peptide-MHC class I interaction. NetMHCpan-2.0 has been trained on the hitherto largest set of quantitative MHC binding data available, covering HLA-A and HLA-B, as well as chimpanzee, rhesus macaque, gorilla, and mouse MHC class I molecules. We show that the NetMHCpan-2.0 method can accurately predict binding to uncharacterized HLA molecules, including HLA-C and HLA-G. Moreover, NetMHCpan-2.0 is demonstrated to accurately predict peptide binding to chimpanzee and macaque MHC class I molecules. The power of NetMHCpan-2.0 to guide immunologists in interpreting cellular immune responses in large out-bred populations is demonstrated. Further, we used NetMHCpan-2.0 to predict potential binding peptides for the pig MHC class I molecule SLA-1*0401. Ninety-three percent of the predicted peptides were demonstrated to bind stronger than 500 nM. The high performance of NetMHCpan-2.0 for non-human primates documents the method's ability to provide broad allelic coverage also beyond human MHC molecules. The method is available at http://www.cbs.dtu.dk/services/NetMHCpan.
Figures




References
-
- Brusic V, Rudy G, Harrison LC. Prediction of MHC binding peptides using artificial neural networks. In: Stonier RJ a. Y. X., editor. Complex systems: mechanism of adaptation. IOS; Amsterdam: 1994. pp. 253–260.
-
- Buus S, Lauemoller SL, Worning P, Kesmir C, Frimurer T, Corbet S, Fomsgaard A, Hilden J, Holm A, Brunak S. Sensitive quantitative predictions of peptide–MHC binding by a ‘Query by Committee’ artificial neural network approach. Tissue Antigens. 2003;62:378–384. doi:10.1034/j.1399-0039.2003.00112.x. - PubMed
-
- Clements CS, Kjer-Nielsen L, McCluskey J, Rossjohn J. Structural studies on HLA-G: implications for ligand and receptor binding. Hum Immunol. 2007;68:220–226. doi:10.1016/j.humimm.2006.09.003. - PubMed
-
- Diehl M, Munz C, Keilholz W, Stevanovic S, Holmes N, Loke YW, Rammensee HG. Nonclassical HLA-G molecules are classical peptide presenters. Curr Biol. 1996;6:305–314. doi:10.1016/S0960-9822(02)00481-5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

