Identification of transgene integration loci of different highly expressing recombinant CHO cell lines by FISH
- PMID: 19002887
- PMCID: PMC3449811
- DOI: 10.1007/s10616-006-9029-0
Identification of transgene integration loci of different highly expressing recombinant CHO cell lines by FISH
Abstract
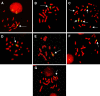
Recombinant CHO cell lines have integrated the expression vectors in various parts of the genome leading to different levels of gene amplification, productivity and stability of protein expression. Identification of insertion sites where gene amplification is possible and the transcription rate is high may lead to systems of site-directed integration and will significantly reduce the process for the generation of stably and highly expressing recombinant cell lines. We have investigated a broad range of recombinant cell lines by FISH analysis and Giemsa-Trypsin banding and analysed their integration loci with regard to the extent of methotrexate pressure, transfection methods, promoters and protein productivities. To summarise, we found that the majority of our high producing recombinant CHO cell lines had integrated the expression construct on a larger chromosome of the genome. Furthermore, except from two cell lines, the exogene was integrated at a single site. The dhfr selection marker was co-localised to the target gene.
Figures




References
-
- Barnett RS, Limoli KL, Huynh TB, Ople EA, Reff ME. Antibody production in Chinese Hamster ovary cells using an impaired selectable marker. In: Wang HY, Imanaka T, editors. Antibody expression and engineering. Washington, DC: American Central Society; 1995.
LinkOut - more resources
Full Text Sources
Other Literature Sources

