Pooling and PCR as a method to combat low frequency gene targeting in mouse embryonic stem cells
- PMID: 19002898
- PMCID: PMC3449676
- DOI: 10.1007/s10616-006-9021-8
Pooling and PCR as a method to combat low frequency gene targeting in mouse embryonic stem cells
Abstract
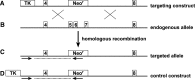
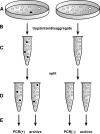
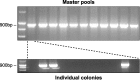
The introduction of germ line modifications by gene targeting in mouse embryonic stem (ES) cells has proven a fundamental technology to relate genes to mammalian biology. Critical aspects required for successful gene targeting have traditionally been experimental enhancements that increase the frequency or detection of homologous recombination within ES cells; however, the utilization of such methods may still result in the failed isolation of a positively targeted ES cell clone. In this study, we discuss the current enhancement methods and describe an ES cell pooling strategy that maximizes the ability to detect properly targeted ES cells regardless of an inherent low targeting efficiency. The sensitivity required to detect correctly targeted events out of a pool of ES cell clones is provided by polymerase chain reaction (PCR), and only those pools containing positives need to be expanded and screened to find individually targeted clones. This method made it possible to identify targeted clones from a screen of approximately 2,300 ES cell colonies by performing only 123 PCR reactions. This technically streamlined approach bypasses the need to troubleshoot and re-engineer an existing targeting construct that is functionally suitable despite its low targeting frequency.
Figures


Similar articles
-
Gene targeting and site-specific recombination in mouse ES cells.Methods Enzymol. 2013;533:133-55. doi: 10.1016/B978-0-12-420067-8.00009-X. Methods Enzymol. 2013. PMID: 24182921
-
Generation of knockout alleles by RFLP based BAC targeting of polymorphic embryonic stem cells.Methods Mol Biol. 2015;1227:143-80. doi: 10.1007/978-1-4939-1652-8_7. Methods Mol Biol. 2015. PMID: 25239745
-
The loss-of-allele assay for ES cell screening and mouse genotyping.Methods Enzymol. 2010;476:295-307. doi: 10.1016/S0076-6879(10)76017-1. Methods Enzymol. 2010. PMID: 20691873
-
Genome engineering via homologous recombination in mouse embryonic stem (ES) cells: an amazingly versatile tool for the study of mammalian biology.An Acad Bras Cienc. 2001 Sep;73(3):365-83. doi: 10.1590/s0001-37652001000300007. An Acad Bras Cienc. 2001. PMID: 11600898 Review.
-
Modifying the mouse: design and desire.Biotechnology (N Y). 1992 May;10(5):534-9. doi: 10.1038/nbt0592-534. Biotechnology (N Y). 1992. PMID: 1368233 Review.
References
-
- DeChiara TM. Gene targeting in ES cells. Methods Mol Biol. 2001;158:19–45. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

