Mechanical stability and differentially conserved physical-chemical properties of titin Ig-domains
- PMID: 19003986
- PMCID: PMC2670939
- DOI: 10.1002/prot.22281
Mechanical stability and differentially conserved physical-chemical properties of titin Ig-domains
Abstract
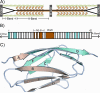
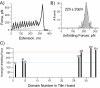
The mechanisms that determine mechanical stabilities of protein folds remain elusive. Our understanding of these mechanisms is vital to both bioengineering efforts and to the better understanding and eventual treatment of pathogenic mutations affecting mechanically important proteins such as titin. We present a new approach to analyze data from single-molecule force spectroscopy for different domains of the giant muscle protein titin. The region of titin found in the I-band of a sarcomere is composed of about 40 Ig-domains and is exposed to force under normal physiological conditions and connects the free-hanging ends of the myosin filaments to the Z-disc. Recent single-molecule force spectroscopy data show a mechanical hierarchy in the I-band domains. Domains near the C-terminus in this region unfold at forces two to three times greater than domains near the beginning of the I-band. Though all of these Ig-domains are thought to share a fold and topology common to members of the Ig-like fold family, the sequences of neighboring domains vary greatly with an average sequence identity of only 25%. We examine in this study the relation of these unique mechanical stabilities of each I-band Ig domain to specific, conserved physical-chemical properties of amino acid sequences in related Ig domains. We find that the sequences of each individual titin Ig domain are very highly conserved, with an average sequence identity of 79% across species that are divergent as humans, chickens, and zebra fish. This indicates that the mechanical properties of each domain are well conserved and tailored to its unique position in the titin molecule. We used the PCPMer software to determine the conservation of amino acid properties in titin Ig domains grouped by unfolding forces into "strong" and "weak" families. We found two motifs unique to each family that may have some role in determining the mechanical properties of these Ig domains. A detailed statistical analysis of properties of individual residues revealed several positions that displayed differentially conserved properties in strong and weak families. In contrast to previous studies, we find evidence that suggests that the mechanical stability of Ig domains is determined by several residues scattered across the beta-sandwich fold, and force sensitive residues are not only confined to the A'-G region.
Figures





Similar articles
-
Structural evidence for a possible role of reversible disulphide bridge formation in the elasticity of the muscle protein titin.Structure. 2001 Apr 4;9(4):331-40. doi: 10.1016/s0969-2126(01)00591-3. Structure. 2001. PMID: 11525170
-
Poly-Ig tandems from I-band titin share extended domain arrangements irrespective of the distinct features of their modular constituents.J Muscle Res Cell Motil. 2005;26(6-8):355-65. doi: 10.1007/s10974-005-9017-6. J Muscle Res Cell Motil. 2005. PMID: 16341830
-
The mechanical stability of immunoglobulin and fibronectin III domains in the muscle protein titin measured by atomic force microscopy.Biophys J. 1998 Dec;75(6):3008-14. doi: 10.1016/S0006-3495(98)77741-0. Biophys J. 1998. PMID: 9826620 Free PMC article.
-
Stretching molecular springs: elasticity of titin filaments in vertebrate striated muscle.Histol Histopathol. 2000 Jul;15(3):799-811. doi: 10.14670/HH-15.799. Histol Histopathol. 2000. PMID: 10963124 Review.
-
Titin elasticity in the context of the sarcomere: force and extensibility measurements on single myofibrils.Adv Exp Med Biol. 2000;481:179-202; discussion 203-6. doi: 10.1007/978-1-4615-4267-4_11. Adv Exp Med Biol. 2000. PMID: 10987073 Review.
Cited by
-
Naturally occurring mutations alter the stability of polycystin-1 polycystic kidney disease (PKD) domains.J Biol Chem. 2009 Nov 20;284(47):32942-9. doi: 10.1074/jbc.M109.021832. Epub 2009 Sep 15. J Biol Chem. 2009. PMID: 19759016 Free PMC article.
-
Structure of giant muscle proteins.Front Physiol. 2013 Dec 12;4:368. doi: 10.3389/fphys.2013.00368. Front Physiol. 2013. PMID: 24376425 Free PMC article. Review.
-
Functional classification of protein toxins as a basis for bioinformatic screening.Sci Rep. 2017 Oct 24;7(1):13940. doi: 10.1038/s41598-017-13957-1. Sci Rep. 2017. PMID: 29066768 Free PMC article.
-
Two immunoglobulin tandem proteins with a linking β-strand reveal unexpected differences in cooperativity and folding pathways.J Mol Biol. 2012 Feb 10;416(1):137-47. doi: 10.1016/j.jmb.2011.12.012. Epub 2011 Dec 13. J Mol Biol. 2012. PMID: 22197372 Free PMC article.
-
Discovery through the computational microscope.Structure. 2009 Oct 14;17(10):1295-306. doi: 10.1016/j.str.2009.09.001. Structure. 2009. PMID: 19836330 Free PMC article. Review.
References
-
- Lange S, Ehler E, Gautel M. From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 2006;16(1):11–18. - PubMed
-
- Linke WA. Sense and stretchability: the role of titin and titin-associated proteins in myocardial stress-sensing and mechanical dysfunction. Cardiovasc Res. 2008;77(4):637–648. - PubMed
-
- Linke WA, Grutzner A. Pulling single molecules of titin by AFM-recent advances and physiological implications. Pflugers Arch. 2007 - PubMed
-
- Tskhovrebova L, Trinick J. Properties of titin immunoglobulin and fibronectin-3 domains. J Biol Chem. 2004;279(45):46351–46354. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

