Comparison of the folding mechanism of highly homologous proteins in the lipid-binding protein family
- PMID: 19003989
- PMCID: PMC6336386
- DOI: 10.1002/prot.22286
Comparison of the folding mechanism of highly homologous proteins in the lipid-binding protein family
Abstract
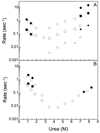
The folding mechanism of two closely related proteins in the intracellular lipid-binding protein family, human bile acid-binding protein (hBABP), and rat bile acid-binding protein (rBABP) were examined. These proteins are 77% identical (93% similar) in sequence. Both of these single domain proteins fit well to a two-state model for unfolding by fluorescence and circular dichroism at equilibrium. Three phases were observed during the unfolding of rBABP by fluorescence but only one phase was observed during the unfolding of hBABP, suggesting that at least two kinetic intermediates accumulate during the unfolding of rBABP that are not observed during the unfolding of hBABP. Fluorine NMR was used to examine the equilibrium unfolding behavior of the W49 side chain in 6-fluorotryptophan-labeled rBABP and hBABP. The structure of rBABP appears to be more dynamic than that of hBABP in the vicinity of W49 in the absence of denaturant, and urea has a greater effect on this dynamic behavior for rBABP than for hBABP. As such, the folding behavior of highly sequence related proteins in this family can be quite different. These differences imply that moderately sized proteins with high sequence and structural similarity can still populate quite different structures during folding.
Copyright 2008 Wiley-Liss, Inc.
Figures






References
-
- Gunasekaran K, Eyles SJ, Hagler AT, Gierasch LM. Keeping it in the family: folding studies of related proteins. Curr Opin Struct Biol. 2001;11(1):83–93. - PubMed
-
- Banaszak L, Winter N, Xu Z, Bernlohr DA, Cowan S, Jones TA. Lipid-binding proteins: A family of fatty acid and retinoid transport proteins. Adv Protein Chem. 1994;45:89–151. - PubMed
-
- Hertzel AV, Bernlohr DA. The mammalian fatty acid-binding protein multigene family: Molecular and genetic insights into function. Trends Endocrinol Metab. 2000;11(5):175–180. - PubMed
-
- Dalessio PM, Ropson IJ. β-sheet proteins with nearly identical structures have different folding intermediates. Biochemistry. 2000;39(5):860–871. - PubMed
-
- Burns LL, Dalessio PM, Ropson IJ. Folding mechanism of three structurally similar β-sheet proteins. Proteins. 1998;33(1):107–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

