DNA barcoding insect-host plant associations
- PMID: 19004756
- PMCID: PMC2660938
- DOI: 10.1098/rspb.2008.1264
DNA barcoding insect-host plant associations
Abstract
Short-sequence fragments ('DNA barcodes') used widely for plant identification and inventorying remain to be applied to complex biological problems. Host-herbivore interactions are fundamental to coevolutionary relationships of a large proportion of species on the Earth, but their study is frequently hampered by limited or unreliable host records. Here we demonstrate that DNA barcodes can greatly improve this situation as they (i) provide a secure identification of host plant species and (ii) establish the authenticity of the trophic association. Host plants of leaf beetles (subfamily Chrysomelinae) from Australia were identified using the chloroplast trnL(UAA) intron as barcodes amplified from beetle DNA extracts. Sequence similarity and phylogenetic analyses provided precise identifications of each host species at tribal, generic and specific levels, depending on the available database coverage in various plant lineages. The 76 species of Chrysomelinae included-more than 10 per cent of the known Australian fauna-feed on 13 plant families, with preference for Australian radiations of Myrtaceae (eucalypts) and Fabaceae (acacias). Phylogenetic analysis of beetles shows general conservation of host association but with rare host shifts between distant plant lineages, including a few cases where barcodes supported two phylogenetically distant host plants. The study demonstrates that plant barcoding is already feasible with the current publicly available data. By sequencing plant barcodes directly from DNA extractions made from herbivorous beetles, strong physical evidence for the host association is provided. Thus, molecular identification using short DNA fragments brings together the detection of species and the analysis of their interactions.
Figures

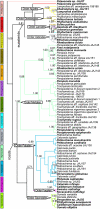

References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi:10.1006/jmbi.1990.9999 - DOI - PubMed
-
- Bradley B.J., Stiller M., Doran-Sheehy D.M., Harris T., Chapman C.A., Vigilant L., Poinar H. Plant DNA sequences from feces: potential means for assessing diets of wild primates. Am. J. Primatol. 2007;69:699–705. doi:10.1002/ajp.20384 - DOI - PubMed
-
- Chase M.W., et al. A proposal for a standardised protocol to barcode all land plants. Taxon. 2007;56:295–299.
-
- Chen Y., Giles K.L., Payton M.E., Greenstone M.H. Identifying key cereal aphid predators by molecular gut analysis. Mol. Ecol. 2000;9:1887–1898. doi:10.1046/j.1365-294x.2000.01100.x - DOI - PubMed
-
- Crisp M., Cook L., Steane D. Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell us about the evolution of diversity in present-day communities? Phil. Trans. R. Soc. B. 2004;359:1551–1571. doi:10.1098/rstb.2004.1528 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
