Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion
- PMID: 19004788
- PMCID: PMC2584702
- DOI: 10.1073/pnas.0807142105
Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion
Abstract
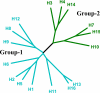
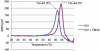
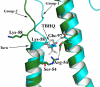
The influenza surface glycoprotein hemagglutinin (HA) is a potential target for antiviral drugs because of its key roles in the initial stages of infection: receptor binding and the fusion of virus and cell membranes. The structure of HA in complex with a known inhibitor of membrane fusion and virus infectivity, tert-butyl hydroquinone (TBHQ), shows that the inhibitor binds in a hydrophobic pocket formed at an interface between HA monomers. Occupation of this site by TBHQ stabilizes the neutral pH structure through intersubunit and intrasubunit interactions that presumably inhibit the conformational rearrangements required for membrane fusion. The nature of the binding site suggests routes for the chemical modification of TBHQ that could lead to the development of more potent inhibitors of membrane fusion and potential anti-influenza drugs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
How influenza virus is locked out of the cell.Proc Natl Acad Sci U S A. 2008 Dec 2;105(48):18647-8. doi: 10.1073/pnas.0810508106. Epub 2008 Nov 24. Proc Natl Acad Sci U S A. 2008. PMID: 19033206 Free PMC article. No abstract available.
References
-
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. - PubMed
-
- Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. 2006;281:8997–9000. - PubMed
-
- Murphy BR, Webster RG. In: Fields Virology. 3rd Ed. Fields DBN, Knipe M, Howley PM, editors. Philadelphia: Lippincott–Raven; 1996. pp. 1397–1445.
-
- Von Itzstein M, et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

