The roles of individual domains of RNase R in substrate binding and exoribonuclease activity. The nuclease domain is sufficient for digestion of structured RNA
- PMID: 19004832
- PMCID: PMC2610503
- DOI: 10.1074/jbc.M806468200
The roles of individual domains of RNase R in substrate binding and exoribonuclease activity. The nuclease domain is sufficient for digestion of structured RNA
Abstract
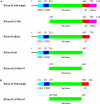
RNase R and RNase II are the two representatives from the RNR family of processive, 3' to 5' exoribonucleases in Escherichia coli. Although RNase II is specific for single-stranded RNA, RNase R readily degrades through structured RNA. Furthermore, RNase R appears to be the only known 3' to 5' exoribonuclease that is able to degrade through double-stranded RNA without the aid of a helicase activity. Consequently, its functional domains and mechanism of action are of great interest. Using a series of truncated RNase R proteins we show that the cold-shock and S1 domains contribute to substrate binding. The cold-shock domains appear to play a role in substrate recruitment, whereas the S1 domain is most likely required to position substrates for efficient catalysis. Most importantly, the nuclease domain alone, devoid of the cold-shock and S1 domains, is sufficient for RNase R to bind and degrade structured RNAs. Moreover, this is a unique property of the nuclease domain of RNase R because this domain in RNase II stalls as it approaches a duplex. We also show that the nuclease domain of RNase R binds RNA more tightly than the nuclease domain of RNase II. This tighter binding may help to explain the difference in catalytic properties between RNase R and RNase II.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

