TGF-beta signals regulate axonal development through distinct Smad-independent mechanisms
- PMID: 19004854
- PMCID: PMC2628568
- DOI: 10.1242/dev.028209
TGF-beta signals regulate axonal development through distinct Smad-independent mechanisms
Abstract
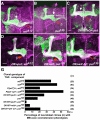
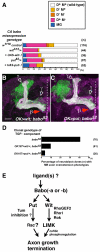
Proper nerve connections form when growing axons terminate at the correct postsynaptic target. Here I show that Transforming growth factor beta (TGFbeta) signals regulate axon growth. In most contexts, TGFbeta signals are tightly linked to Smad transcriptional activity. Although known to exist, how Smad-independent pathways mediate TGFbeta responses in vivo is unclear. In Drosophila mushroom body (MB) neurons, loss of the TGFbeta receptor Baboon (Babo) results in axon overextension. Conversely, misexpression of constitutively active Babo results in premature axon termination. Smad activity is not required for these phenotypes. This study shows that Babo signals require the Rho GTPases Rho1 and Rac, and LIM kinase1 (LIMK1), which regulate the actin cytoskeleton. Contrary to the well-established receptor activation model, in which type 1 receptors act downstream of type 2 receptors, this study shows that the type 2 receptors Wishful thinking (Wit) and Punt act downstream of the Babo type 1 receptor. Wit and Punt regulate axon growth independently, and interchangeably, through LIMK1-dependent and -independent mechanisms. Thus, novel TGFbeta receptor interactions control non-Smad signals and regulate multiple aspects of axonal development in vivo.
Figures






Similar articles
-
Visualization of Endogenous Type I TGF-β Receptor Baboon in the Drosophila Brain.Sci Rep. 2020 Mar 20;10(1):5132. doi: 10.1038/s41598-020-61950-y. Sci Rep. 2020. PMID: 32198477 Free PMC article.
-
Rho GTPases regulate axon growth through convergent and divergent signaling pathways.Neuron. 2004 Dec 2;44(5):779-93. doi: 10.1016/j.neuron.2004.11.014. Neuron. 2004. PMID: 15572110
-
The NAV2 homolog Sickie regulates F-actin-mediated axonal growth in Drosophila mushroom body neurons via the non-canonical Rac-Cofilin pathway.Development. 2014 Dec;141(24):4716-28. doi: 10.1242/dev.113308. Epub 2014 Nov 19. Development. 2014. PMID: 25411210
-
TGF-beta family signal transduction in Drosophila development: from Mad to Smads.Dev Biol. 1999 Jun 15;210(2):251-68. doi: 10.1006/dbio.1999.9282. Dev Biol. 1999. PMID: 10357889 Review.
-
Rac proteins and the control of axon development.Curr Opin Neurobiol. 2003 Jun;13(3):384-90. doi: 10.1016/s0959-4388(03)00071-0. Curr Opin Neurobiol. 2003. PMID: 12850224 Review.
Cited by
-
Parallel Activin and BMP signaling coordinates R7/R8 photoreceptor subtype pairing in the stochastic Drosophila retina.Elife. 2017 Aug 30;6:e25301. doi: 10.7554/eLife.25301. Elife. 2017. PMID: 28853393 Free PMC article.
-
Lipophorin receptors regulate mushroom body development and complex behaviors in Drosophila.BMC Biol. 2022 Sep 7;20(1):198. doi: 10.1186/s12915-022-01393-1. BMC Biol. 2022. PMID: 36071487 Free PMC article.
-
Smad3 regulates Rho signaling via NET1 in the transforming growth factor-beta-induced epithelial-mesenchymal transition of human retinal pigment epithelial cells.J Biol Chem. 2010 Aug 20;285(34):26618-27. doi: 10.1074/jbc.M109.073155. Epub 2010 Jun 11. J Biol Chem. 2010. PMID: 20547485 Free PMC article.
-
Drosophila CORL is required for Smad2-mediated activation of Ecdysone Receptor expression in the mushroom body.Development. 2012 Sep;139(18):3392-401. doi: 10.1242/dev.079442. Epub 2012 Aug 8. Development. 2012. PMID: 22874913 Free PMC article.
-
R-Smad competition controls activin receptor output in Drosophila.PLoS One. 2012;7(5):e36548. doi: 10.1371/journal.pone.0036548. Epub 2012 May 1. PLoS One. 2012. PMID: 22563507 Free PMC article.
References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–95. - PubMed
-
- Augsburger A, Schuchardt A, Hoskins S, Dodd J, Butler S. BMPs as mediators of roof plate repulsion of commissural neurons. Neuron. 1999;24:127–41. - PubMed
-
- Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

