Pre-P is a secreted glycoprotein encoded as an N-terminal extension of the duck hepatitis B virus polymerase gene
- PMID: 19004940
- PMCID: PMC2620893
- DOI: 10.1128/JVI.01263-08
Pre-P is a secreted glycoprotein encoded as an N-terminal extension of the duck hepatitis B virus polymerase gene
Abstract
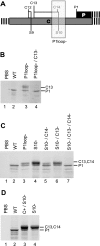
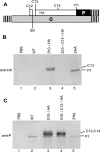
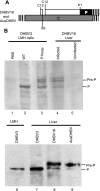
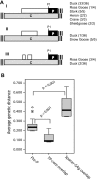
The duck hepatitis B virus (DHBV) pregenomic RNA is a bicistronic mRNA encoding the core and polymerase proteins. Thirteen AUGs (C2 to C14) and 10 stop codons (S1 to S10) are located between the C1 AUG for the core protein and the P1 AUG that initiates polymerase translation. We previously found that the translation of the DHBV polymerase is initiated by ribosomal shunting. Here, we assessed the biosynthetic events after shunting. Translation of the polymerase open reading frame was found to initiate at the C13, C14, and P1 AUGs. Initiation at the C13 AUG occurred through ribosomal shunting because translation from this codon was cap dependent but was insensitive to blocking ribosomal scanning internally in the message. C13 and C14 are in frame with P1, and translation from these upstream start codons led to the production of larger isoforms of P. We named these isoforms "pre-P" by analogy to the pre-C and pre-S regions of the core and surface antigen open reading frames. Pre-P was produced in DHBV16 and AusDHBV-infected duck liver and was predicted to exist in 80% of avian hepadnavirus strains. Pre-P was not encapsidated into DHBV core particles, and the viable strain DHBV3 cannot make pre-P, so it is not essential for viral replication. Surprisingly, we found that pre-P is an N-linked glycoprotein that is secreted into the medium of cultured cells. These data indicate that DHBV produces an additional protein that has not been previously reported. Identifying the role of pre-P may improve our understanding of the biology of DHBV infection.
Figures




References
-
- Beck, J., H. Bartos, and M. Nassal. 1997. Experimental confirmation of a hepatitis B (HBV) epsilon-like bulge and loop structure in avian HBV RNA encapsidation signals. Virology 227500-504. - PubMed
-
- Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 2942376-2378. - PubMed
-
- Cao, F., and J. E. Tavis. 2006. Suppression of mRNA accumulation by the duck hepatitis B virus reverse transcriptase. Virology 350475-483. - PubMed
-
- Chang, S. F., H. J. Netter, M. Bruns, R. Schneider, K. Frolich, and H. Will. 1999. A new avian hepadnavirus infecting snow geese (Anser caerulescens) produces a significant fraction of virions containing single-stranded DNA. Virology 26239-54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

