Effects of simultaneous deletion of pUL11 and glycoprotein M on virion maturation of herpes simplex virus type 1
- PMID: 19004941
- PMCID: PMC2612385
- DOI: 10.1128/JVI.01842-08
Effects of simultaneous deletion of pUL11 and glycoprotein M on virion maturation of herpes simplex virus type 1
Abstract
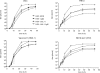
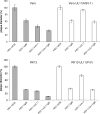
The conserved membrane-associated tegument protein pUL11 and envelope glycoprotein M (gM) are involved in secondary envelopment of herpesvirus nucleocapsids in the cytoplasm. Although deletion of either gene had only moderate effects on replication of the related alphaherpesviruses herpes simplex virus type 1 (HSV-1) and pseudorabies virus (PrV) in cell culture, simultaneous deletion of both genes resulted in a severe impairment in virion morphogenesis of PrV coinciding with the formation of huge inclusions in the cytoplasm containing nucleocapsids embedded in tegument (M. Kopp, H. Granzow, W. Fuchs, B. G. Klupp, and T. C. Mettenleiter, J. Virol. 78:3024-3034, 2004). To test whether a similar phenotype occurs in HSV-1, a gM and pUL11 double deletion mutant was generated based on a newly established bacterial artificial chromosome clone of HSV-1 strain KOS. Since gM-negative HSV-1 has not been thoroughly investigated ultrastructurally and different phenotypes have been ascribed to pUL11-negative HSV-1, single gene deletion mutants were also constructed and analyzed. On monkey kidney (Vero) cells, deletion of either pUL11 or gM resulted in ca.-fivefold-reduced titers and 40- to 50%-reduced plaque diameters compared to those of wild-type HSV-1 KOS, while on rabbit kidney (RK13) cells the defects were more pronounced, resulting in ca.-50-fold titer and 70% plaque size reduction for either mutant. Electron microscopy revealed that in the absence of either pUL11 or gM virion formation in the cytoplasm was inhibited, whereas nuclear stages were not visibly affected, which is in line with the phenotypes of corresponding PrV mutants. Simultaneous deletion of pUL11 and gM led to additive growth defects and, in RK13 cells, to the formation of large intracytoplasmic inclusions of capsids and tegument material, comparable to those in PrV-DeltaUL11/gM-infected RK13 cells. The defects of HSV-1DeltaUL11 and HSV-1DeltaUL11/gM could be partially corrected in trans by pUL11 of PrV. Thus, our data indicate that PrV and HSV-1 pUL11 and gM exhibit similar functions in cytoplasmic steps of virion assembly.
Figures






Similar articles
-
Phenotypic similarities and differences between UL37-deleted pseudorabies virus and herpes simplex virus type 1.J Gen Virol. 2009 Jul;90(Pt 7):1560-1568. doi: 10.1099/vir.0.010322-0. Epub 2009 Mar 18. J Gen Virol. 2009. PMID: 19297610
-
Partial functional complementation of a pseudorabies virus UL25 deletion mutant by herpes simplex virus type 1 pUL25 indicates overlapping functions of alphaherpesvirus pUL25 proteins.J Virol. 2008 Jun;82(12):5725-34. doi: 10.1128/JVI.02441-07. Epub 2008 Apr 9. J Virol. 2008. PMID: 18400859 Free PMC article.
-
Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus.J Virol. 1999 Jul;73(7):5364-72. doi: 10.1128/JVI.73.7.5364-5372.1999. J Virol. 1999. PMID: 10364283 Free PMC article.
-
Tegument Assembly and Secondary Envelopment of Alphaherpesviruses.Viruses. 2015 Sep 18;7(9):5084-114. doi: 10.3390/v7092861. Viruses. 2015. PMID: 26393641 Free PMC article. Review.
-
Involvement of Terminase Complex in Herpes Simplex Virus Mature Virion Egress.Curr Protein Pept Sci. 2022;23(2):105-113. doi: 10.2174/1389203723666220217144432. Curr Protein Pept Sci. 2022. PMID: 35176987 Review.
Cited by
-
The Roles of Envelope Glycoprotein M in the Life Cycle of Some Alphaherpesviruses.Front Microbiol. 2021 Feb 19;12:631523. doi: 10.3389/fmicb.2021.631523. eCollection 2021. Front Microbiol. 2021. PMID: 33679658 Free PMC article. Review.
-
Sequence variability in clinical and laboratory isolates of herpes simplex virus 1 reveals new mutations.J Virol. 2010 May;84(10):5303-13. doi: 10.1128/JVI.00312-10. Epub 2010 Mar 10. J Virol. 2010. PMID: 20219902 Free PMC article.
-
Characterization of pseudorabies virus (PrV) cleavage-encapsidation proteins and functional complementation of PrV pUL32 by the homologous protein of herpes simplex virus type 1.J Virol. 2009 Apr;83(8):3930-43. doi: 10.1128/JVI.02636-08. Epub 2009 Feb 4. J Virol. 2009. PMID: 19193798 Free PMC article.
-
N-Linked Glycosylation and Expression of Duck Plague Virus pUL10 Promoted by pUL49.5.Microbiol Spectr. 2023 Aug 17;11(4):e0162523. doi: 10.1128/spectrum.01625-23. Epub 2023 Jun 28. Microbiol Spectr. 2023. PMID: 37378543 Free PMC article.
-
Glycoprotein D of HSV-1 is dependent on tegument protein UL16 for packaging and contains a motif that is differentially required for syncytia formation.Virology. 2019 Jan 15;527:64-76. doi: 10.1016/j.virol.2018.09.018. Epub 2018 Nov 19. Virology. 2019. PMID: 30465930 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

