A single amino acid substitution in a segment of the CA protein within Gag that has similarity to human immunodeficiency virus type 1 blocks infectivity of a human endogenous retrovirus K provirus in the human genome
- PMID: 19004950
- PMCID: PMC2612375
- DOI: 10.1128/JVI.01439-08
A single amino acid substitution in a segment of the CA protein within Gag that has similarity to human immunodeficiency virus type 1 blocks infectivity of a human endogenous retrovirus K provirus in the human genome
Abstract
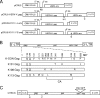
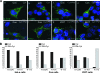
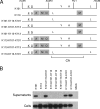
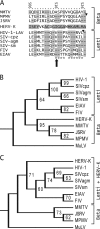
Human endogenous retrovirus K (HERV-K) is the most intact retrovirus in the human genome. However, no single HERV-K provirus in the human genome today appears to be infectious. Since the Gag protein is the central component for the production of retrovirus particles, we investigated the abilities of Gag from two HERV-K proviruses to support production of virus-like particles and viral infectivity. HERV-K113 has full-length open reading frames for all viral proteins, while HERV-K101 has a full-length gag open reading frame and is expressed in human male germ cell tumors. The Gag of HERV-K101 allowed production of viral particles and infectivity, although at lower levels than observed with a consensus sequence Gag. Thus, including HERV-K109, at least two HERV-K proviruses in human genome today have functional Gag proteins. In contrast, HERV-K113 Gag supported only very low levels of particle production, and no infectivity was detectable due to a single amino acid substitution (I516M) near the extreme C terminus of the CA protein within Gag. The sequence of this portion of HERV-K CA showed similarities to that of human immunodeficiency virus type 1 and other primate immunodeficiency viruses. The extreme C terminus of CA may be a general determinant of retrovirus particle production. In addition, precise mapping of the defects in HERV-K proviruses as was done here identifies the key polymorphisms that need to be analyzed to assess the possible existence of infectious HERV-K alleles within the human population.
Figures



Similar articles
-
Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles.J Gen Virol. 2008 Feb;89(Pt 2):567-572. doi: 10.1099/vir.0.83534-0. J Gen Virol. 2008. PMID: 18198388
-
Insertional polymorphisms of full-length endogenous retroviruses in humans.Curr Biol. 2001 Oct 2;11(19):1531-5. doi: 10.1016/s0960-9822(01)00455-9. Curr Biol. 2001. PMID: 11591322
-
Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K.J Virol. 1999 Nov;73(11):9187-95. doi: 10.1128/JVI.73.11.9187-9195.1999. J Virol. 1999. PMID: 10516026 Free PMC article.
-
Molecular biology of type A endogenous retrovirus.Kitasato Arch Exp Med. 1990 Sep;63(2-3):77-90. Kitasato Arch Exp Med. 1990. PMID: 1710682 Review.
-
HERV-K: the biologically most active human endogenous retrovirus family.J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13 Suppl 1:S261-7. doi: 10.1097/00042560-199600001-00039. J Acquir Immune Defic Syndr Hum Retrovirol. 1996. PMID: 8797733 Review.
Cited by
-
Reconstitution of the ancestral glycoprotein of human endogenous retrovirus k and modulation of its functional activity by truncation of the cytoplasmic domain.J Virol. 2009 Dec;83(24):12790-800. doi: 10.1128/JVI.01368-09. Epub 2009 Oct 7. J Virol. 2009. PMID: 19812154 Free PMC article.
-
Variant splicing and influence of ionizing radiation on human endogenous retrovirus K (HERV-K) transcripts in cancer cell lines.PLoS One. 2013 Oct 18;8(10):e76472. doi: 10.1371/journal.pone.0076472. eCollection 2013. PLoS One. 2013. PMID: 24204631 Free PMC article.
-
Downregulation of Human Endogenous Retrovirus Type K (HERV-K) Viral env RNA in Pancreatic Cancer Cells Decreases Cell Proliferation and Tumor Growth.Clin Cancer Res. 2017 Oct 1;23(19):5892-5911. doi: 10.1158/1078-0432.CCR-17-0001. Epub 2017 Jul 5. Clin Cancer Res. 2017. PMID: 28679769 Free PMC article.
-
HERV-K HML-2 diversity among humans.Proc Natl Acad Sci U S A. 2016 Apr 19;113(16):4240-2. doi: 10.1073/pnas.1603569113. Epub 2016 Apr 8. Proc Natl Acad Sci U S A. 2016. PMID: 27071126 Free PMC article. No abstract available.
-
Molecular mechanisms by which HERV-K Gag interferes with HIV-1 Gag assembly and particle infectivity.Retrovirology. 2017 Apr 26;14(1):27. doi: 10.1186/s12977-017-0351-8. Retrovirology. 2017. PMID: 28446240 Free PMC article.
References
-
- Adamson, C. S., and E. O. Freed. 2007. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv. Pharmacol. 55347-387. - PubMed
-
- Armbruester, V., M. Sauter, E. Krautkraemer, E. Meese, A. Kleiman, B. Best, K. Roemer, and N. Mueller-Lantzsch. 2002. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin. Cancer Res. 81800-1807. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

