The immediate early gene arc/arg3.1: regulation, mechanisms, and function
- PMID: 19005037
- PMCID: PMC2615463
- DOI: 10.1523/JNEUROSCI.3864-08.2008
The immediate early gene arc/arg3.1: regulation, mechanisms, and function
Abstract
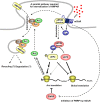
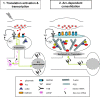
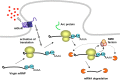
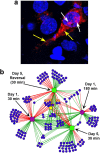
In a manner unique among activity-regulated immediate early genes (IEGs), mRNA encoded by Arc (also known as Arg3.1) undergoes rapid transport to dendrites and local synaptic translation. Despite this intrinsic appeal, relatively little is known about the neuronal and behavioral functions of Arc or its molecular mechanisms of action. Here, we attempt to distill recent advances on Arc spanning its transcriptional and translational regulation, the functions of the Arc protein in multiple forms of neuronal plasticity [long-term potentiation (LTP), long-term depression (LTD), and homeostatic plasticity], and its broader role in neural networks of behaving animals. Worley and colleagues have shown that Arc interacts with endophilin and dynamin, creating a postsynaptic trafficking endosome that selectively modifies the expression of AMPA-type glutamate receptors at the excitatory synapses. Both LTD and homeostatic plasticity in the hippocampus are critically dependent on Arc-mediated endocytosis of AMPA receptors. LTD evoked by activation of metabotropic glutamate receptors depends on rapid Arc translation controlled by elongation factor 2. Bramham and colleagues have shown that sustained translation of newly induced Arc mRNA is necessary for cofilin phosphorylation and stable expansion of the F-actin cytoskeleton underlying LTP consolidation in the dentate gyrus of live rats. In addition to regulating F-actin, Arc synthesis maintains the activity of key translation factors during LTP consolidation. This process of Arc-dependent consolidation is activated by the secretory neurotrophin, BDNF. Moore and colleagues have shown that Arc mRNA is a natural target for nonsense-mediated mRNA decay (NMD) by virtue of its two conserved 3'-UTR introns. NMD and other related translation-dependent mRNA decay mechanisms may serve as critical brakes on protein expression that contribute to the fine spatial-temporal control of Arc synthesis. In studies in behaving rats, Guzowski and colleagues have shown that location-specific firing of CA3 and CA1 hippocampal neurons in the presence of theta rhythm provides the necessary stimuli for activation of Arc transcription. The impact of Arc transcription in memory processes may depend on the specific context of coexpressed IEGs, in addition to posttranscriptional regulation of Arc by neuromodulatory inputs from the amygdala and other brain regions. In sum, Arc is emerging as a versatile, finely tuned system capable of coupling changes in neuronal activity patterns to diverse forms of synaptic plasticity, thereby optimizing information storage in active networks.
Figures
References
-
- Bingol B, Schuman EM. Synaptic protein degradation by the ubiquitin proteasome system. Curr Opin Neurobiol. 2005;15:536–541. - PubMed
-
- Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:1144–1148. - PubMed
-
- Bloomer WA, VanDongen HM, VanDongen AM. Activity-regulated cytoskeleton-associated protein Arc/Arg3.1 binds to spectrin and associates with nuclear promyelocytic leukemia (PML) bodies. Brain Res. 2007;1153:20–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
