Confirmation of rubella within 4 days of rash onset: comparison of rubella virus RNA detection in oral fluid with immunoglobulin M detection in serum or oral fluid
- PMID: 19005151
- PMCID: PMC2620850
- DOI: 10.1128/JCM.01231-08
Confirmation of rubella within 4 days of rash onset: comparison of rubella virus RNA detection in oral fluid with immunoglobulin M detection in serum or oral fluid
Abstract
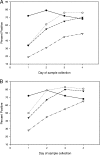
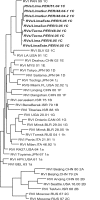
Rubella virus infection is typically diagnosed by the identification of rubella virus-specific immunoglobulin M (IgM) antibodies in serum, but approximately 50% of serum samples from rubella cases collected on the day of rash onset are negative for rubella virus-specific IgM. The ability to detect IgM in sera and oral fluids was compared with the ability to detect rubella virus RNA in oral fluids by reverse transcription-PCR (RT-PCR) by using paired samples taken within the first 4 days after rash onset from suspected rubella cases during an outbreak in Perú. Sera were tested for IgM by both indirect and capture enzyme immunoassays (EIAs), and oral fluids were tested for IgM by a capture EIA. Tests for IgM in serum were more sensitive for the confirmation of rubella than the test for IgM in oral fluid during the 4 days after rash onset. RT-PCR confirmed more suspected cases than serum IgM tests on days 1 and 2 after rash onset. The methods confirmed approximately the same number of cases on days 3 and 4 after rash onset. However, a few cases were detected by serum IgM tests but not by RT-PCR even on the day of rash onset. Nine RT-PCR-positive oral fluid specimens were shown to contain rubella virus sequences of genotype 1C. In summary, RT-PCR testing of oral fluid confirmed more rubella cases than IgM testing of either serum or oral fluid samples collected in the first 2 days after rash onset; the maximum number of confirmations of rubella cases was obtained by combining RT-PCR and serology testing.
Figures

References
-
- Bellini, W. J., and J. P. Icenogle. 2007. Measles and rubella viruses, p. 1378-1391. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
-
- Best, J. M., and S. O'Shea. 1995. Rubella virus, p. 583-600. In E. H. Lennette, D. A. Lennette, and E. T. Lennette, (ed.) Diagnostic procedures for viral, rickettsial, and chlamydial infections. American Public Health Association, Washington, DC.
-
- Centers for Disease Control and Prevention. 2005. Global measles and rubella laboratory network, January 2004-June 2005. MMWR Morb. Mortal. Wkly. Rep. 541100-1104. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

