Anc1, a protein associated with multiple transcription complexes, is involved in postreplication repair pathway in S. cerevisiae
- PMID: 19005567
- PMCID: PMC2579579
- DOI: 10.1371/journal.pone.0003717
Anc1, a protein associated with multiple transcription complexes, is involved in postreplication repair pathway in S. cerevisiae
Abstract
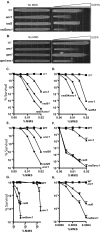
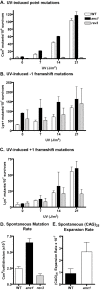
Yeast strains lacking Anc1, a member of the YEATS protein family, are sensitive to several DNA damaging agents. The YEATS family includes two human genes that are common fusion partners with MLL in human acute leukemias. Anc1 is a member of seven multi-protein complexes involved in transcription, and the damage sensitivity observed in anc1Delta cells is mirrored in strains deleted for some other non-essential members of several of these complexes. Here we show that ANC1 is in the same epistasis group as SRS2 and RAD5, members of the postreplication repair (PRR) pathway, but has additive or synergistic interactions with several other members of this pathway. Although PRR is traditionally divided into an "error-prone" and an "error-free" branch, ANC1 is not epistatic with all members of either established branch, and instead defines a new error-free branch of the PRR pathway. Like several genes involved in PRR, an intact ANC1 gene significantly suppresses spontaneous mutation rates, including the expansion of (CAG)(25) repeats. Anc1's role in the PRR pathway, as well as its role in suppressing triplet repeats, point to a possible mechanism for a protein of potential medical interest.
Conflict of interest statement
Figures

References
-
- Begley TJ, Rosenbach AS, Ideker T, Samson LD. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol Cancer Res. 2002;1:103–112. - PubMed
-
- Begley TJ, Rosenbach AS, Ideker T, Samson LD. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Mol Cell. 2004;16:117–125. - PubMed
-
- Henry NL, Campbell AM, Feaver WJ, Poon D, Weil PA, et al. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 1994;8:2868–2878. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

