Cavities and atomic packing in protein structures and interfaces
- PMID: 19005575
- PMCID: PMC2582456
- DOI: 10.1371/journal.pcbi.1000188
Cavities and atomic packing in protein structures and interfaces
Abstract
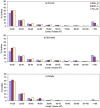
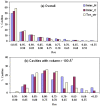
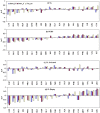
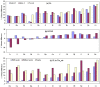
A comparative analysis of cavities enclosed in a tertiary structure of proteins and interfaces formed by the interaction of two protein subunits in obligate and non-obligate categories (represented by homodimeric molecules and heterocomplexes, respectively) is presented. The total volume of cavities increases with the size of the protein (or the interface), though the exact relationship may vary in different cases. Likewise, for individual cavities also there is quantitative dependence of the volume on the number of atoms (or residues) lining the cavity. The larger cavities tend to be less spherical, solvated, and the interfaces are enriched in these. On average 15 A(3) of cavity volume is found to accommodate single water, with another 40-45 A(3) needed for each additional solvent molecule. Polar atoms/residues have a higher propensity to line solvated cavities. Relative to the frequency of occurrence in the whole structure (or interface), residues in beta-strands are found more often lining the cavities, and those in turn and loop the least. Any depression in one chain not complemented by a protrusion in the other results in a cavity in the protein-protein interface. Through the use of the Voronoi volume, the packing of residues involved in protein-protein interaction has been compared to that in the protein interior. For a comparable number of atoms the interface has about twice the number of cavities relative to the tertiary structure.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- Richards FM. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. - PubMed
-
- Richards FM. The interpretation of protein structures: total volume, group volume distributions and packing density. J Mol Biol. 1974;82:1–14. - PubMed
-
- Gerstein M, Sonnhammer EL, Chothia C. Volume changes in protein evolution. J Mol Biol. 1994;236:1067–1078. - PubMed
-
- Fleming PJ, Richards FM. Protein packing: dependence on protein size, secondary structure and amino acid composition. J Mol Biol. 2000;299:487–498. - PubMed
-
- Connolly ML. Atomic size packing defects in proteins. Int J Pept Protein Res. 1986;28:360–363. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

