Potential intra- and intermolecular interactions involving the unique-5' region of the HIV-1 5'-UTR
- PMID: 19006324
- PMCID: PMC2646082
- DOI: 10.1021/bi8014373
Potential intra- and intermolecular interactions involving the unique-5' region of the HIV-1 5'-UTR
Abstract
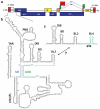
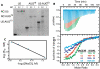
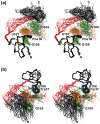
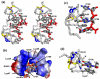
The 5'-untranslated region (5'-UTR) of the human immunodeficiency virus type-1 (HIV-1) genome regulates multiple RNA-dependent functions during viral replication and has been proposed to adopt multiple secondary structures. Recent phylogenetic studies identified base pair complementarity between residues of the unique 5' element and those near the gag start codon (gag(AUG)) that is conserved among evolutionarily distant retroviruses, suggesting a potential long-range RNA-RNA interaction. However, nucleotide accessibility studies led to conflicting conclusions about the presence of such interactions in virions and in infected cells. Here, we show that an 11-nucleotide oligo-RNA spanning residues 105-115 of the 5'-UTR (U5) readily binds to oligoribonucleotides containing the gag start codon (AUG), disrupting a pre-existing stem loop and forming a heteroduplex stabilized by 11 Watson-Crick base pairs (K(d) = 0.47 +/- 0.16 microM). Addition of the HIV-1 nucleocapsid protein (NC), the trans-acting viral factor required for genome packaging, disrupts the heteroduplex by binding tightly to U5 (K(d) = 122 +/- 10 nM). The structure of the NC:U5 complex, determined by NMR, exhibits features similar to those observed in NC complexes with HIV-1 stem loop RNAs, including the insertion of guanosine nucleobases to hydrophobic clefts on the surface of the zinc fingers and a 3'-to-5' orientation of the RNA relative to protein. Our findings indicate that the previously proposed long-range U5-gag(AUG) interaction is feasible and suggest a potential NC-dependent mechanism for modulating the structure of the 5'-UTR.
Figures


Similar articles
-
Conserved determinants of lentiviral genome dimerization.Retrovirology. 2015 Sep 29;12:83. doi: 10.1186/s12977-015-0209-x. Retrovirology. 2015. PMID: 26420212 Free PMC article.
-
Unpaired Guanosines in the 5' Untranslated Region of HIV-1 RNA Act Synergistically To Mediate Genome Packaging.J Virol. 2020 Oct 14;94(21):e00439-20. doi: 10.1128/JVI.00439-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32796062 Free PMC article.
-
Role of the 5' TAR stem--loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA.Biochemistry. 2008 Mar 11;47(10):3283-93. doi: 10.1021/bi7023173. Epub 2008 Feb 16. Biochemistry. 2008. PMID: 18278873
-
NMR detection of structures in the HIV-1 5'-leader RNA that regulate genome packaging.Science. 2011 Oct 14;334(6053):242-5. doi: 10.1126/science.1210460. Science. 2011. PMID: 21998393 Free PMC article.
-
Structural determinants and mechanism of HIV-1 genome packaging.J Mol Biol. 2011 Jul 22;410(4):609-33. doi: 10.1016/j.jmb.2011.04.029. J Mol Biol. 2011. PMID: 21762803 Free PMC article. Review.
Cited by
-
Intrinsic nucleic acid dynamics modulates HIV-1 nucleocapsid protein binding to its targets.PLoS One. 2012;7(6):e38905. doi: 10.1371/journal.pone.0038905. Epub 2012 Jun 20. PLoS One. 2012. PMID: 22745685 Free PMC article.
-
Retrospective on the all-in-one retroviral nucleocapsid protein.Virus Res. 2014 Nov 26;193:2-15. doi: 10.1016/j.virusres.2014.05.011. Epub 2014 Jun 4. Virus Res. 2014. PMID: 24907482 Free PMC article. Review.
-
Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes.J Mol Biol. 2009 Dec 4;394(3):544-57. doi: 10.1016/j.jmb.2009.09.040. Epub 2009 Sep 24. J Mol Biol. 2009. PMID: 19782089 Free PMC article.
-
Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal.Proc Natl Acad Sci U S A. 2020 Jul 28;117(30):17737-17746. doi: 10.1073/pnas.2008519117. Epub 2020 Jul 9. Proc Natl Acad Sci U S A. 2020. PMID: 32647061 Free PMC article.
-
A structure-based mechanism for tRNA and retroviral RNA remodelling during primer annealing.Nature. 2014 Nov 27;515(7528):591-5. doi: 10.1038/nature13709. Epub 2014 Sep 7. Nature. 2014. PMID: 25209668
References
-
- Berkhout B. Prog. Nucl. Acid Res. and Mol. Biol. Academic Press, Inc.; 1996. Structure and function of the human immunodeficiency virus leader RNA; pp. 1–34. - PubMed
-
- Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr. Top. Microbiol. Immun. 1996;214:177–218. - PubMed
-
- Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, N.Y.: 1997. - PubMed
-
- Swanstrom R, Wills JW. Synthesis, assembly and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Press; Plainview, N.Y.: 1997. pp. 263–334. - PubMed
-
- D'Souza V, Summers MF. How retroviruses select their genomes. Nature Reviews Microbiology. 2005;3:643–655. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

