Molecular analysis of DMP1 mutants causing autosomal recessive hypophosphatemic rickets
- PMID: 19007919
- PMCID: PMC2669955
- DOI: 10.1016/j.bone.2008.10.040
Molecular analysis of DMP1 mutants causing autosomal recessive hypophosphatemic rickets
Abstract
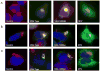
We previously demonstrated that the mutations Met1Val (M1V) and the deletion of nucleotides 1484-1490 (1484-1490del) in Dentin matrix protein-1 (DMP1) cause the novel disorder autosomal recessive hypophosphatemic rickets (ARHR), which is associated with elevated fibroblast growth factor-23 (FGF23). To further understand the role of DMP1 in ARHR, we undertook molecular genetic and in vitro expression studies. First, we examined a kindred with a severe hypophosphatemic rickets phenotype and recessive inheritance. Analyses of this family demonstrated that the affected members had elevated serum FGF23 and carried a large, biallelic deletion that removed the majority of DMP1. At a minimum, this deletion encompassed 49 kb between DMP1 exon 3 and an intergenic region 5' to the next telomeric gene, integrin-binding sialoprotein (IBSP). We next performed immunofluorescent studies in cells to understand the effects of the known ARHR mutations on DMP1 cellular processing. These analyses showed that the M1V DMP1 mutant was not sorted to the trans-Golgi network (TGN) and secretory pathway, but filled the entire cytoplasm. In contrast, the 1484-1490del mutant localized to the TGN and was secreted, similar to wild type DMP1. The 1484-1490del mutation replaces the DMP1 18 C-terminal amino acids with 33 non-native residues. Truncation of wild type DMP1 by these native 18 residues followed by Western blot and confocal microscopic analyses demonstrated a wild type expression pattern when compared with the 1484-1490del mutant, indicating that the last 18 residues are not critical for cellular trafficking, but that the 33 additional residues arising from the 1484-1490del mutation likely compromise DMP1 processing. The relationship between DMP1 and FGF23 is unclear. To test endogenous DMP1 response to serum metabolites that also regulate FGF23, UMR-106 cells were treated with 1,25(OH)(2) vitamin D (1x10(-7) M) and showed a 12-fold increase in DMP1 mRNA and protein at 24 h. In summary, we have identified a novel DMP1 deletion as the cause of ARHR, as well as demonstrated that the ARHR mutations alter DMP1 cellular processing, and that DMP1 can be regulated by vitamin D. Taken together, this work expands our understanding of the genetic and molecular mechanisms associated with DMP1 alterations causing ARHR.
Figures




References
-
- Lorenz-Depiereux B, Bastepe M, Benet-Pages A, Amyere M, Wagenstaller J, Muller-Barth U, Badenhoop K, Kaiser SM, Rittmaster RS, Shlossberg AH, Olivares JL, Loris C, Ramos FJ, Glorieux F, Vikkula M, Juppner H, Strom TM. DMP1 mutations in autosomal recessive hypophosphatemia implicate a bone matrix protein in the regulation of phosphate homeostasis. Nat Genet. 2006;38:1248–50. - PMC - PubMed
-
- Fisher LW, Fedarko NS. Six genes expressed in bones and teeth encode the current members of the SIBLING family of proteins. Connect Tissue Res. 2003;44(Suppl 1):33–40. - PubMed
-
- Butler WT, Ritchie H. The nature and functional significance of dentin extracellular matrix proteins. Int J Dev Biol. 1995;39:169–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

