Calcium-dependent binding of HCN1 channel protein to hair cell stereociliary tip link protein protocadherin 15 CD3
- PMID: 19008224
- PMCID: PMC2631981
- DOI: 10.1074/jbc.M806177200
Calcium-dependent binding of HCN1 channel protein to hair cell stereociliary tip link protein protocadherin 15 CD3
Abstract
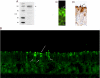
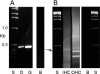
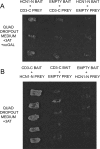
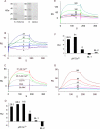
The cytoplasmic amino terminus of HCN1, the primary full-length HCN isoform expressed in trout saccular hair cells, was found by yeast two-hybrid protocols to bind the cytoplasmic carboxyl-terminal domain of a protocadherin 15a-like protein. HCN1 was immunolocalized to discrete sites on saccular hair cell stereocilia, consistent with gradated distribution expected for tip link sites of protocadherin 15a. HCN1 message was also detected in cDNA libraries of rat cochlear inner and outer hair cells, and HCN1 protein was immunolocalized to cochlear hair cell stereocilia. As predicted by the trout hair cell model, the amino terminus of rat organ of Corti HCN1 was found by yeast two-hybrid analysis to bind the carboxyl terminus of protocadherin 15 CD3, a tip link protein implicated in mechanosensory transduction. Specific binding between HCN1 and protocadherin 15 CD3 was confirmed with pull-down assays and surface plasmon resonance analysis, both predicting dependence on Ca(2+). In the presence of calcium chelators, binding between HCN1 and protocadherin 15 CD3 was characterized by a K(D) = 2.39 x 10(-7) m. Ca(2+) at 26.5-68.0 microm promoted binding, with K(D) = 5.26 x 10(-8) m (at 61 microm Ca(2+)). Binding by deletion mutants of protocadherin 15 CD3 pointed to amino acids 158-179 (GenBank accession number XP_238200), with homology to the comparable region in trout hair cell protocadherin 15a-like protein, as necessary for binding to HCN1. Amino terminus binding of HCN1 to HCN1, hypothesized to underlie HCN1 channel formation, was also found to be Ca(2+)-dependent, although the binding was skewed toward a lower effective maximum [Ca(2+)] than for the HCN1 interaction with protocadherin 15 CD3. Competition may therefore exist in vivo between the two binding sites for HCN1, with binding of HCN1 to protocadherin 15 CD3 favored between 26.5 and 68 microm Ca(2+). Taken together, the evidence supports a role for HCN1 in mechanosensory transduction of inner ear hair cells.
Figures




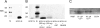

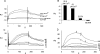
References
-
- Cho, W. J., Drescher, M. J., Hatfield, J. S., Bessert, D. A., Skoff, R. P., and Drescher, D. G. (2003) Neuroscience 118 525-534 - PubMed
-
- Holt, J. R., and Eatock, R. A. (1995) J. Neurophysiol. 73 1484-1502 - PubMed
-
- Ramakrishnan, N. A., Drescher, M. J., and Drescher, D. G. (2007) Assoc. Res. Otolaryngol. Abstr. 30 31
-
- Proenza, C., Angoli, D., Agranovich, E., Macri, V., and Accili, E. A. (2002) J. Biol. Chem. 277 5101-5109 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

