Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1
- PMID: 19008854
- PMCID: PMC2609736
- DOI: 10.1038/emboj.2008.241
Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1
Abstract
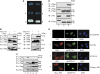
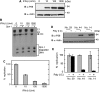
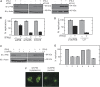
Type I interferon (IFN) inhibits, by an unknown mechanism, the replication of human papillomaviruses (HPV), which are major human pathogens, Here, we present evidence that P56 (a protein), the expression of which is strongly induced by IFN, double-stranded RNA and viruses, mediates the anti-HPV effect of IFN. Ectopic expression of P56 inhibited HPV DNA replication and its ablation in IFN-treated cells alleviated the inhibitory effect of IFN on HPV DNA replication. Protein-protein interaction and mutational analyses established that the antiviral effect of P56 was mediated by its direct interaction with the DNA replication origin-binding protein E1 of several strains of HPV, through the tetratricopeptide repeat 2 in the N-terminal region of P56 and the C-terminal region of E1. In vivo, the interaction with P56, a cytoplasmic protein, caused translocation of E1 from the nucleus to the cytoplasm. In vitro, recombinant P56, or a small fragment derived from it, inhibited the DNA helicase activity of E1 and E1-mediated HPV DNA replication. These observations delineate the molecular mechanism of IFN's antiviral action against HPV.
Figures




Similar articles
-
The inhibitory action of P56 on select functions of E1 mediates interferon's effect on human papillomavirus DNA replication.J Virol. 2010 Dec;84(24):13036-9. doi: 10.1128/JVI.01194-10. Epub 2010 Oct 6. J Virol. 2010. PMID: 20926571 Free PMC article.
-
E1-mediated recruitment of a UAF1-USP deubiquitinase complex facilitates human papillomavirus DNA replication.J Virol. 2014 Aug;88(15):8545-55. doi: 10.1128/JVI.00379-14. Epub 2014 May 21. J Virol. 2014. PMID: 24850727 Free PMC article.
-
Artificial Recruitment of UAF1-USP Complexes by a PHLPP1-E1 Chimeric Helicase Enhances Human Papillomavirus DNA Replication.J Virol. 2015 Jun;89(12):6227-39. doi: 10.1128/JVI.00560-15. Epub 2015 Apr 1. J Virol. 2015. PMID: 25833051 Free PMC article.
-
Human papillomaviruses and the interferon response.J Interferon Cytokine Res. 2009 Sep;29(9):629-35. doi: 10.1089/jir.2009.0075. J Interferon Cytokine Res. 2009. PMID: 19715460 Free PMC article. Review.
-
The E1 proteins.Virology. 2013 Oct;445(1-2):35-56. doi: 10.1016/j.virol.2013.07.020. Epub 2013 Sep 10. Virology. 2013. PMID: 24029589 Free PMC article. Review.
Cited by
-
Antiviral innate immunity disturbs podocyte cell function.J Innate Immun. 2013;5(3):231-41. doi: 10.1159/000345255. Epub 2012 Dec 22. J Innate Immun. 2013. PMID: 23296190 Free PMC article.
-
Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals.Elife. 2016 May 31;5:e14228. doi: 10.7554/eLife.14228. Elife. 2016. PMID: 27240734 Free PMC article.
-
Epigallocatechin gallate inhibits cell growth and regulates miRNA expression in cervical carcinoma cell lines infected with different high-risk human papillomavirus subtypes.Exp Ther Med. 2019 Mar;17(3):1742-1748. doi: 10.3892/etm.2018.7131. Epub 2018 Dec 24. Exp Ther Med. 2019. PMID: 30783443 Free PMC article.
-
IFIT5 Participates in the Antiviral Mechanisms of Rainbow Trout Red Blood Cells.Front Immunol. 2019 Apr 16;10:613. doi: 10.3389/fimmu.2019.00613. eCollection 2019. Front Immunol. 2019. PMID: 31040842 Free PMC article.
-
Regulation of the life cycle of HPVs by differentiation and the DNA damage response.Future Microbiol. 2013 Dec;8(12):1547-57. doi: 10.2217/fmb.13.127. Future Microbiol. 2013. PMID: 24266355 Free PMC article. Review.
References
-
- Asano K, Merrick WC, Hershey JW (1997) The translation initiation factor eIF3-p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J Biol Chem 272: 23477–23480 - PubMed
-
- Auster AS, Joshua-Tor L (2004) The DNA-binding domain of human papillomavirus type 18 E1. Crystal structure, dimerization, and DNA binding. J Biol Chem 279: 3733–3742 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

