Silencing and transcriptional properties of the imprinted Airn ncRNA are independent of the endogenous promoter
- PMID: 19008856
- PMCID: PMC2599881
- DOI: 10.1038/emboj.2008.239
Silencing and transcriptional properties of the imprinted Airn ncRNA are independent of the endogenous promoter
Abstract
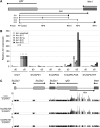
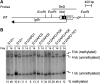
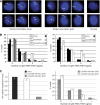
The Airn macro ncRNA is the master regulator of imprinted expression in the Igf2r imprinted gene cluster where it silences three flanking genes in cis. Airn transcription shows unusual features normally viewed as promoter specific, such as impaired post-transcriptional processing and a macro size. The Airn transcript is 108 kb long, predominantly unspliced and nuclear localized, with only a minority being variably spliced and exported. Here, we show by deletion of the Airn ncRNA promoter and replacement with a constitutive strong or weak promoter that splicing suppression and termination, as well as silencing activity, are maintained by strong Airn expression from an exogenous promoter. This indicates that all functional regions are located within the Airn transcript. DNA methylation of the maternal imprint control element (ICE) restricts Airn expression to the paternal allele and we also show that a strong active promoter is required to maintain the unmethylated state of the paternal ICE. Thus, Airn expression not only induces silencing of flanking mRNA genes but also protects the paternal copy of the ICE from de novo methylation.
Figures




Similar articles
-
A downstream CpG island controls transcript initiation and elongation and the methylation state of the imprinted Airn macro ncRNA promoter.PLoS Genet. 2012;8(3):e1002540. doi: 10.1371/journal.pgen.1002540. Epub 2012 Mar 1. PLoS Genet. 2012. PMID: 22396659 Free PMC article.
-
Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing.Science. 2012 Dec 14;338(6113):1469-72. doi: 10.1126/science.1228110. Science. 2012. PMID: 23239737
-
Imprinted Igf2r silencing depends on continuous Airn lncRNA expression and is not restricted to a developmental window.Development. 2013 Mar;140(6):1184-95. doi: 10.1242/dev.088849. Development. 2013. PMID: 23444351
-
Kcnq1ot1: a chromatin regulatory RNA.Semin Cell Dev Biol. 2011 Jun;22(4):343-50. doi: 10.1016/j.semcdb.2011.02.020. Epub 2011 Feb 21. Semin Cell Dev Biol. 2011. PMID: 21345374 Review.
-
Macro lncRNAs: a new layer of cis-regulatory information in the mammalian genome.RNA Biol. 2012 Jun;9(6):731-41. doi: 10.4161/rna.19985. Epub 2012 May 23. RNA Biol. 2012. PMID: 22617879 Review.
Cited by
-
The function of non-coding RNAs in genomic imprinting.Development. 2009 Jun;136(11):1771-83. doi: 10.1242/dev.030403. Development. 2009. PMID: 19429783 Free PMC article. Review.
-
Evolution of the unspliced transcriptome.BMC Evol Biol. 2015 Aug 20;15:166. doi: 10.1186/s12862-015-0437-7. BMC Evol Biol. 2015. PMID: 26289325 Free PMC article.
-
Maternal transmission of a humanised Igf2r allele results in an Igf2 dependent hypomorphic and non-viable growth phenotype.PLoS One. 2013;8(2):e57270. doi: 10.1371/journal.pone.0057270. Epub 2013 Feb 28. PLoS One. 2013. PMID: 23468951 Free PMC article.
-
Comparative RNA Genomics.Methods Mol Biol. 2024;2802:347-393. doi: 10.1007/978-1-0716-3838-5_12. Methods Mol Biol. 2024. PMID: 38819565
-
Maternal transmission of an Igf2r domain 11: IGF2 binding mutant allele (Igf2rI1565A) results in partial lethality, overgrowth and intestinal adenoma progression.Sci Rep. 2019 Aug 6;9(1):11388. doi: 10.1038/s41598-019-47827-9. Sci Rep. 2019. PMID: 31388182 Free PMC article.
References
-
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R (2006) Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44: 23–28 - PubMed
-
- Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E (2007) mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett 581: 2845–2853 - PubMed
-
- Bentley DL (2005) Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17: 251–256 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

