Quantification of local morphodynamics and local GTPase activity by edge evolution tracking
- PMID: 19008941
- PMCID: PMC2573959
- DOI: 10.1371/journal.pcbi.1000223
Quantification of local morphodynamics and local GTPase activity by edge evolution tracking
Abstract
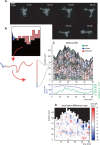
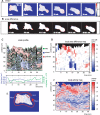
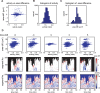
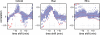
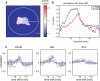
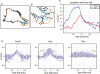
Advances in time-lapse fluorescence microscopy have enabled us to directly observe dynamic cellular phenomena. Although the techniques themselves have promoted the understanding of dynamic cellular functions, the vast number of images acquired has generated a need for automated processing tools to extract statistical information. A problem underlying the analysis of time-lapse cell images is the lack of rigorous methods to extract morphodynamic properties. Here, we propose an algorithm called edge evolution tracking (EET) to quantify the relationship between local morphological changes and local fluorescence intensities around a cell edge using time-lapse microscopy images. This algorithm enables us to trace the local edge extension and contraction by defining subdivided edges and their corresponding positions in successive frames. Thus, this algorithm enables the investigation of cross-correlations between local morphological changes and local intensity of fluorescent signals by considering the time shifts. By applying EET to fluorescence resonance energy transfer images of the Rho-family GTPases Rac1, Cdc42, and RhoA, we examined the cross-correlation between the local area difference and GTPase activity. The calculated correlations changed with time-shifts as expected, but surprisingly, the peak of the correlation coefficients appeared with a 6-8 min time shift of morphological changes and preceded the Rac1 or Cdc42 activities. Our method enables the quantification of the dynamics of local morphological change and local protein activity and statistical investigation of the relationship between them by considering time shifts in the relationship. Thus, this algorithm extends the value of time-lapse imaging data to better understand dynamics of cellular function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
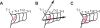
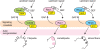

References
-
- Lecuit T, Lenne PF. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol. 2007;8:633–644. - PubMed
-
- Bakal C, Aach J, Church G, Perrimon N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 2007;316:1753–1756. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

