Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative
- PMID: 19011064
- PMCID: PMC2620721
- DOI: 10.1128/AEM.01037-08
Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative
Abstract
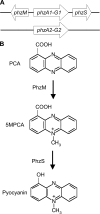
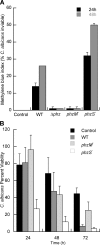
Phenazines are redox-active small molecules that play significant roles in the interactions between pseudomonads and diverse eukaryotes, including fungi. When Pseudomonas aeruginosa and Candida albicans were cocultured on solid medium, a red pigmentation developed that was dependent on P. aeruginosa phenazine biosynthetic genes. Through a genetic screen in combination with biochemical experiments, it was found that a P. aeruginosa-produced precursor to pyocyanin, proposed to be 5-methyl-phenazinium-1-carboxylate (5MPCA), was necessary for the formation of the red pigmentation. The 5MPCA-derived pigment was found to accumulate exclusively within fungal cells, where it retained the ability to be reversibly oxidized and reduced, and its detection correlated with decreased fungal viability. Pyocyanin was not required for pigment formation or fungal killing. Spectral analyses showed that the partially purified pigment from within the fungus differed from aeruginosins A and B, two red phenazine derivatives formed late in P. aeruginosa cultures. The red pigment isolated from C. albicans that had been cocultured with P. aeruginosa was heterogeneous and difficult to release from fungal cells, suggesting its modification within the fungus. These findings suggest that intracellular targeting of some phenazines may contribute to their toxicity and that this strategy could be useful in developing new antifungals.
Figures



References
-
- Bauernfeind, A., R. M. Bertele, K. Harms, G. Horl, R. Jungwirth, C. Petermuller, B. Przyklenk, and C. Weisslein-Pfister. 1987. Qualitative and quantitative microbiological analysis of sputa of 102 patients with cystic fibrosis. Infection 15:270-277. - PubMed
-
- Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

