Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions
- PMID: 19011103
- PMCID: PMC2587588
- DOI: 10.1073/pnas.0806759105
Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions
Abstract
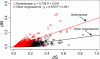
Angiosperm plastid genomes are generally conserved in gene content and order with rates of nucleotide substitutions for protein-coding genes lower than for nuclear protein-coding genes. A few groups have experienced genomic change, and extreme changes in gene content and order are found within the flowering plant family Geraniaceae. The complete plastid genome sequence of Pelargonium X hortorum (Geraniaceae) reveals the largest and most rearranged plastid genome identified to date. Highly elevated rates of sequence evolution in Geraniaceae mitochondrial genomes have been reported, but rates in Geraniaceae plastid genomes have not been characterized. Analysis of nucleotide substitution rates for 72 plastid genes for 47 angiosperm taxa, including nine Geraniaceae, show that values of dN are highly accelerated in ribosomal protein and RNA polymerase genes throughout the family. Furthermore, dN/dS is significantly elevated in the same two classes of plastid genes as well as in ATPase genes. A relatively high dN/dS ratio could be interpreted as evidence of two phenomena, namely positive or relaxed selection, neither of which is consistent with our current understanding of plastid genome evolution in photosynthetic plants. These analyses are the first to use protein-coding sequences from complete plastid genomes to characterize rates and patterns of sequence evolution for a broad sampling of photosynthetic angiosperms, and they reveal unprecedented accumulation of nucleotide substitutions in Geraniaceae. To explain these remarkable substitution patterns in the highly rearranged Geraniaceae plastid genomes, we propose a model of aberrant DNA repair coupled with altered gene expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Raubeson LA, Jansen RK. In: Plant Diversity And Evolution: Genotypic And Phenotypic Variation In Higher Plants. Henry RJ, editor. Cambridge, MA: CABI Publishing; 2005. pp. 45–68.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

