Medium- and short-chain dehydrogenase/reductase gene and protein families : the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes
- PMID: 19011750
- PMCID: PMC2792337
- DOI: 10.1007/s00018-008-8588-y
Medium- and short-chain dehydrogenase/reductase gene and protein families : the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes
Abstract
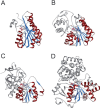
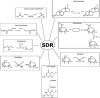
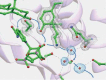
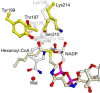
Short-chain dehydrogenases/reductases (SDRs) constitute a large family of NAD(P)(H)-dependent oxidoreductases, sharing sequence motifs and displaying similar mechanisms. SDR enzymes have critical roles in lipid, amino acid, carbohydrate, cofactor, hormone and xenobiotic metabolism as well as in redox sensor mechanisms. Sequence identities are low, and the most conserved feature is an alpha/beta folding pattern with a central beta sheet flanked by 2 - 3 alpha-helices from each side, thus a classical Rossmannfold motif for nucleotide binding. The conservation of this element and an active site, often with an Asn-Ser-Tyr-Lys tetrad, provides a platform for enzymatic activities encompassing several EC classes, including oxidoreductases, epimerases and lyases. The common mechanism is an underlying hydride and proton transfer involving the nicotinamide and typically an active site tyrosine residue, whereas substrate specificity is determined by a variable C-terminal segment. Relationships exist with bacterial haloalcohol dehalogenases, which lack cofactor binding but have the active site architecture, emphasizing the versatility of the basic fold in also generating hydride transfer-independent lyases. The conserved fold and nucleotide binding emphasize the role of SDRs as scaffolds for an NAD(P)(H) redox sensor system, of importance to control metabolic routes, transcription and signalling.
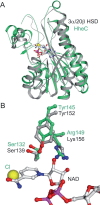
Figures
References
-
- Jörnvall H., Höög J. O., Persson B. SDR and MDR: completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett. 1999;445:261–264. - PubMed
-
- Pearl F., Todd A., Sillitoe I., Dibley M., Redfern O., Lewis T., Bennett C., Marsden R., Grant A., Lee D., et al. The CATH Domain Structure Database and related resources Gene3D and DHS provide comprehensive domain family information for genome analysis. Nucleic Acids Res. 2005;33:D247–251. - PMC - PubMed
-
- Nobel S., Abrahmsen L., Oppermann U. Metabolic conversion as a pre-receptor control mechanism for lipophilic hormones. Eur. J. Biochem. 2001;268:4113–4125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

