Alterations in CDH15 and KIRREL3 in patients with mild to severe intellectual disability
- PMID: 19012874
- PMCID: PMC2668064
- DOI: 10.1016/j.ajhg.2008.10.020
Alterations in CDH15 and KIRREL3 in patients with mild to severe intellectual disability
Abstract
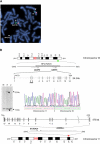
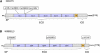
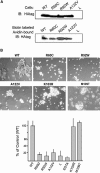
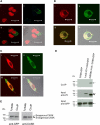
Cell-adhesion molecules play critical roles in brain development, as well as maintaining synaptic structure, function, and plasticity. Here we have found the disruption of two genes encoding putative cell-adhesion molecules, CDH15 (cadherin superfamily) and KIRREL3 (immunoglobulin superfamily), by a chromosomal translocation t(11;16) in a female patient with intellectual disability (ID). We screened coding regions of these two genes in a cohort of patients with ID and controls and identified four nonsynonymous CDH15 variants and three nonsynonymous KIRREL3 variants that appear rare and unique to ID. These variations altered highly conserved residues and were absent in more than 600 unrelated patients with ID and 800 control individuals. Furthermore, in vivo expression studies showed that three of the CDH15 variations adversely altered its ability to mediate cell-cell adhesion. We also show that in neuronal cells, human KIRREL3 colocalizes and interacts with the synaptic scaffolding protein, CASK, recently implicated in X-linked brain malformation and ID. Taken together, our data suggest that alterations in CDH15 and KIRREL3, either alone or in combination with other factors, could play a role in phenotypic expression of ID in some patients.
Figures
References
-
- American Association on Mental Retardation . Tenth Edition. American Association on Mental Retardation; Washington, DC: 2002. Mental Retardation: Definition, Classification and Systems of Support.
-
- Ropers H.H. Genetics of intellectual disability. Curr. Opin. Genet. Dev. 2008;18:241–250. - PubMed
-
- Chelly J., Khelfaoui M., Francis F., Cherif B., Bienvenu T. Genetics and pathophysiology of mental retardation. Eur. J. Hum. Genet. 2006;14:701–713. - PubMed
-
- Chiurazzi P., Schwartz C.E., Gecz J., Neri G. XLMR genes: Update 2007. Eur. J. Hum. Genet. 2008;16:422–434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

