Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death
- PMID: 19013277
- PMCID: PMC2684502
- DOI: 10.1016/j.cell.2008.09.038
Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death
Abstract
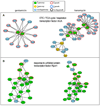
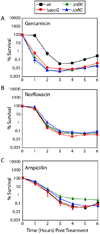
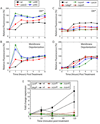
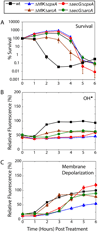
Aminoglycoside antibiotics, such as gentamicin and kanamycin, directly target the ribosome, yet the mechanisms by which these bactericidal drugs induce cell death are not fully understood. Recently, oxidative stress has been implicated as one of the mechanisms whereby bactericidal antibiotics kill bacteria. Here, we use systems-level approaches and phenotypic analyses to provide insight into the pathway whereby aminoglycosides ultimately trigger hydroxyl radical formation. We show, by disabling systems that facilitate membrane protein traffic, that mistranslation and misfolding of membrane proteins are central to aminoglycoside-induced oxidative stress and cell death. Signaling through the envelope stress-response two-component system is found to be a key player in this process, and the redox-responsive two-component system is shown to have an associated role. Additionally, we show that these two-component systems play a general role in bactericidal antibiotic-mediated oxidative stress and cell death, expanding our understanding of the common mechanism of killing induced by bactericidal antibiotics.
Figures



References
-
- Akiyama Y, Kihara A, Tokuda H, Ito K. FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J Biol Chem. 1996;271:31196–31201. - PubMed
-
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

