FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast
- PMID: 19013280
- PMCID: PMC2703716
- DOI: 10.1016/j.cell.2008.09.037
FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast
Abstract
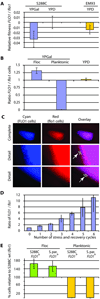
The budding yeast, Saccharomyces cerevisiae, has emerged as an archetype of eukaryotic cell biology. Here we show that S. cerevisiae is also a model for the evolution of cooperative behavior by revisiting flocculation, a self-adherence phenotype lacking in most laboratory strains. Expression of the gene FLO1 in the laboratory strain S288C restores flocculation, an altered physiological state, reminiscent of bacterial biofilms. Flocculation protects the FLO1 expressing cells from multiple stresses, including antimicrobials and ethanol. Furthermore, FLO1(+) cells avoid exploitation by nonexpressing flo1 cells by self/non-self recognition: FLO1(+) cells preferentially stick to one another, regardless of genetic relatedness across the rest of the genome. Flocculation, therefore, is driven by one of a few known "green beard genes," which direct cooperation toward other carriers of the same gene. Moreover, FLO1 is highly variable among strains both in expression and in sequence, suggesting that flocculation in S. cerevisiae is a dynamic, rapidly evolving social trait.
Figures




Comment in
-
A social life for discerning microbes.Cell. 2008 Nov 14;135(4):600-3. doi: 10.1016/j.cell.2008.10.030. Cell. 2008. PMID: 19013271 Review.
References
-
- Arthington-Skaggs BA, Lee-Yang W, Ciblak MA, Frade JP, Brandt ME, Hajjeh RA, Harrison LH, Sofair AN, Warnock DW. Comparison of visual and spectrophotometric methods of broth microdilution MIC end point determination and evaluation of a sterol quantitation method for in vitro susceptibility testing of fluconazole and itraconazole against trailing and nontrailing Candida isolates. Antimicrob Agents Chemother. 2002;46:2477–2481. - PMC - PubMed
-
- Beauvais A, Schmidt C, Guadagnini S, Roux P, Perret E, Henry C, Paris S, Mallet A, Prevost MC, Latge JP. An extracellular matrix glues together the aerial-grown hyphae of Aspergillus fumigatus. Cell Microbiol. 2007;9:1588–1600. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

