Transcription factor expression dynamics of early T-lymphocyte specification and commitment
- PMID: 19013443
- PMCID: PMC2663971
- DOI: 10.1016/j.ydbio.2008.10.021
Transcription factor expression dynamics of early T-lymphocyte specification and commitment
Abstract
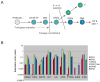
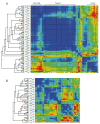
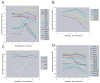
Mammalian T lymphocytes are a prototype for development from adult pluripotent stem cells. While T-cell specification is driven by Notch signaling, T-lineage commitment is only finalized after prolonged Notch activation. However, no T-lineage specific regulatory factor has been reported that mediates commitment. We used a gene-discovery approach to identify additional candidate T-lineage transcription factors and characterized expression of >100 regulatory genes in early T-cell precursors using realtime RT-PCR. These regulatory genes were also monitored in multilineage precursors as they entered T-cell or non-T-cell pathways in vitro; in non-T cells ex vivo; and in later T-cell developmental stages after lineage commitment. At least three major expression patterns were observed. Transcription factors in the largest group are expressed at relatively stable levels throughout T-lineage specification as a legacy from prethymic precursors, with some continuing while others are downregulated after commitment. Another group is highly expressed in the earliest stages only, and is downregulated before or during commitment. Genes in a third group undergo upregulation at one of three distinct transitions, suggesting a positive regulatory cascade. However, the transcription factors induced during commitment are not T-lineage specific. Different members of the same transcription factor family can follow opposite trajectories during specification and commitment, while factors co-expressed early can be expressed in divergent patterns in later T-cell development. Some factors reveal new regulatory distinctions between alphabeta and gammadelta T-lineage differentiation. These results show that T-cell identity has an essentially complex regulatory basis and provide a detailed framework for regulatory network modeling of T-cell specification.
Figures



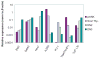

References
-
- Anderson MK. At the crossroads: diverse roles of early thymocyte transcriptional regulators. Immunol Rev. 2006;209:191–211. - PubMed
-
- Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–3148. - PubMed
-
- Anderson MK, Hernandez-Hoyos G, Dionne CJ, Arias A, Chen D, Rothenberg EV. Definition of regulatory network elements for T-cell development by perturbation analysis with PU.1 and GATA-3. Devel Biol. 2002a;246:103–121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

