CellProfiler Analyst: data exploration and analysis software for complex image-based screens
- PMID: 19014601
- PMCID: PMC2614436
- DOI: 10.1186/1471-2105-9-482
CellProfiler Analyst: data exploration and analysis software for complex image-based screens
Abstract
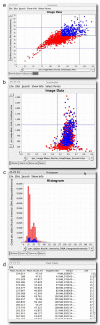
Background: Image-based screens can produce hundreds of measured features for each of hundreds of millions of individual cells in a single experiment.
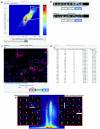
Results: Here, we describe CellProfiler Analyst, open-source software for the interactive exploration and analysis of multidimensional data, particularly data from high-throughput, image-based experiments.
Conclusion: The system enables interactive data exploration for image-based screens and automated scoring of complex phenotypes that require combinations of multiple measured features per cell.
Figures






References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

