Analysis of the Pythium ultimum transcriptome using Sanger and Pyrosequencing approaches
- PMID: 19014603
- PMCID: PMC2612028
- DOI: 10.1186/1471-2164-9-542
Analysis of the Pythium ultimum transcriptome using Sanger and Pyrosequencing approaches
Abstract
Background: Pythium species are an agriculturally important genus of plant pathogens, yet are not understood well at the molecular, genetic, or genomic level. They are closely related to other oomycete plant pathogens such as Phytophthora species and are ubiquitous in their geographic distribution and host rage. To gain a better understanding of its gene complement, we generated Expressed Sequence Tags (ESTs) from the transcriptome of Pythium ultimum DAOM BR144 (= ATCC 200006 = CBS 805.95) using two high throughput sequencing methods, Sanger-based chain termination sequencing and pyrosequencing-based sequencing-by-synthesis.
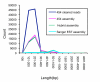
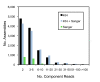
Results: A single half-plate pyrosequencing (454 FLX) run on adapter-ligated cDNA from a normalized cDNA population generated 90,664 reads with an average read length of 190 nucleotides following cleaning and removal of sequences shorter than 100 base pairs. After clustering and assembly, a total of 35,507 unique sequences were generated. In parallel, 9,578 reads were generated from a library constructed from the same normalized cDNA population using dideoxy chain termination Sanger sequencing, which upon clustering and assembly generated 4,689 unique sequences. A hybrid assembly of both Sanger- and pyrosequencing-derived ESTs resulted in 34,495 unique sequences with 1,110 sequences (3.2%) that were solely derived from Sanger sequencing alone. A high degree of similarity was seen between P. ultimum sequences and other sequenced plant pathogenic oomycetes with 91% of the hybrid assembly derived sequences > 500 bp having similarity to sequences from plant pathogenic Phytophthora species. An analysis of Gene Ontology assignments revealed a similar representation of molecular function ontologies in the hybrid assembly in comparison to the predicted proteomes of three Phytophthora species, suggesting a broad representation of the P. ultimum transcriptome was present in the normalized cDNA population. P. ultimum sequences with similarity to oomycete RXLR and Crinkler effectors, Kazal-like and cystatin-like protease inhibitors, and elicitins were identified. Sequences with similarity to thiamine biosynthesis enzymes that are lacking in the genome sequences of three Phytophthora species and one downy mildew were identified and could serve as useful phylogenetic markers. Furthermore, we identified 179 candidate simple sequence repeats that can be used for genotyping strains of P. ultimum.
Conclusion: Through these two technologies, we were able to generate a robust set (approximately 10 Mb) of transcribed sequences for P. ultimum. We were able to identify known sequences present in oomycetes as well as identify novel sequences. An ample number of candidate polymorphic markers were identified in the dataset providing resources for phylogenetic and diagnostic marker development for this species. On a technical level, in spite of the depth possible with 454 FLX platform, the Sanger and pyro-based sequencing methodologies were complementary as each method generated sequences unique to each platform.
Figures

Similar articles
-
Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire.Genome Biol. 2010;11(7):R73. doi: 10.1186/gb-2010-11-7-r73. Epub 2010 Jul 13. Genome Biol. 2010. PMID: 20626842 Free PMC article.
-
Insights into the Host Specificity of a New Oomycete Root Pathogen, Pythium brassicum P1: Whole Genome Sequencing and Comparative Analysis Reveals Contracted Regulation of Metabolism, Protein Families, and Distinct Pathogenicity Repertoire.Int J Mol Sci. 2021 Aug 20;22(16):9002. doi: 10.3390/ijms22169002. Int J Mol Sci. 2021. PMID: 34445718 Free PMC article.
-
Comparative genomics reveals insight into virulence strategies of plant pathogenic oomycetes.PLoS One. 2013 Oct 4;8(10):e75072. doi: 10.1371/journal.pone.0075072. eCollection 2013. PLoS One. 2013. PMID: 24124466 Free PMC article.
-
The Top 10 oomycete pathogens in molecular plant pathology.Mol Plant Pathol. 2015 May;16(4):413-34. doi: 10.1111/mpp.12190. Epub 2014 Dec 11. Mol Plant Pathol. 2015. PMID: 25178392 Free PMC article. Review.
-
Mastering DNA chromatogram analysis in Sanger sequencing for reliable clinical analysis.J Genet Eng Biotechnol. 2023 Nov 13;21(1):115. doi: 10.1186/s43141-023-00587-6. J Genet Eng Biotechnol. 2023. PMID: 37955813 Free PMC article. Review.
Cited by
-
RNA-seq analysis of transcriptome and glucosinolate metabolism in seeds and sprouts of broccoli (Brassica oleracea var. italic).PLoS One. 2014 Feb 27;9(2):e88804. doi: 10.1371/journal.pone.0088804. eCollection 2014. PLoS One. 2014. PMID: 24586398 Free PMC article.
-
De novo sequencing and analysis of the Ulva linza transcriptome to discover putative mechanisms associated with its successful colonization of coastal ecosystems.BMC Genomics. 2012 Oct 25;13:565. doi: 10.1186/1471-2164-13-565. BMC Genomics. 2012. PMID: 23098051 Free PMC article.
-
Subtractive libraries for prospecting differentially expressed genes in the soybean under water deficit.Genet Mol Biol. 2012 Jun;35(1 (suppl)):304-14. doi: 10.1590/S1415-47572012000200011. Genet Mol Biol. 2012. PMID: 22802715 Free PMC article.
-
Transcriptome analysis of green and purple fruited pepper provides insight into novel regulatory genes in anthocyanin biosynthesis.PeerJ. 2024 Jan 17;12:e16792. doi: 10.7717/peerj.16792. eCollection 2024. PeerJ. 2024. PMID: 38250728 Free PMC article.
-
Next-generation sequencing techniques for eukaryotic microorganisms: sequencing-based solutions to biological problems.Eukaryot Cell. 2010 Sep;9(9):1300-10. doi: 10.1128/EC.00123-10. Epub 2010 Jul 2. Eukaryot Cell. 2010. PMID: 20601439 Free PMC article. Review.
References
-
- Index Fungorum http://www.indexfungorum.org
-
- Plaats-Niterink AJ van der. Monograph of the genus Pythium. Studies in Mycology. 1981;21:1–242.
-
- Cook RJ, Sitton JW, Waldher JT. Evidence for Pythium as a pathogen of direct-drilled wheat in the Pacific Northwest. Plant Dis. 1980;64:102–103.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

