Mechanisms for human genomic rearrangements
- PMID: 19014668
- PMCID: PMC2583991
- DOI: 10.1186/1755-8417-1-4
Mechanisms for human genomic rearrangements
Abstract
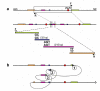
Genomic rearrangements describe gross DNA changes of the size ranging from a couple of hundred base pairs, the size of an average exon, to megabases (Mb). When greater than 3 to 5 Mb, such changes are usually visible microscopically by chromosome studies. Human diseases that result from genomic rearrangements have been called genomic disorders. Three major mechanisms have been proposed for genomic rearrangements in the human genome. Non-allelic homologous recombination (NAHR) is mostly mediated by low-copy repeats (LCRs) with recombination hotspots, gene conversion and apparent minimal efficient processing segments. NAHR accounts for most of the recurrent rearrangements: those that share a common size, show clustering of breakpoints, and recur in multiple individuals. Non-recurrent rearrangements are of different sizes in each patient, but may share a smallest region of overlap whose change in copy number may result in shared clinical features among different patients. LCRs do not mediate, but may stimulate non-recurrent events. Some rare NAHRs can also be mediated by highly homologous repetitive sequences (for example, Alu, LINE); these NAHRs account for some of the non-recurrent rearrangements. Other non-recurrent rearrangements can be explained by non-homologous end-joining (NHEJ) and the Fork Stalling and Template Switching (FoSTeS) models. These mechanisms occur both in germ cells, where the rearrangements can be associated with genomic disorders, and in somatic cells in which such genomic rearrangements can cause disorders such as cancer. NAHR, NHEJ and FoSTeS probably account for the majority of genomic rearrangements in our genome and the frequency distribution of the three at a given locus may partially reflect the genomic architecture in proximity to that locus. We provide a review of the current understanding of these three models.
Figures





References
-
- Lupski JR, Stankiewicz P. Genomic Disorders. Totowa, New Jersey: Humana Press; 2006.
LinkOut - more resources
Full Text Sources
Miscellaneous

