Identification of the Rock-dependent transcriptome in rodent fibroblasts
- PMID: 19015069
- PMCID: PMC2737644
- DOI: 10.1007/s12094-008-0279-5
Identification of the Rock-dependent transcriptome in rodent fibroblasts
Abstract
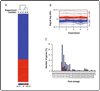
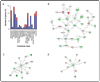
Rock proteins are Rho GTPase-dependent serine/ threonine kinases with crucial roles in F-actin dynamics and cell transformation. By analogy with other protein kinase families, it can be assumed that Rock proteins act, at least in part, through the regulation of gene expression events. However, with the exception of some singular transcriptional targets recently identified, the actual impact of these kinases on the overall cell transcriptome remains unknown. To address this issue, we have used a microarray approach to compare the transcriptomes of exponentially growing NIH3T3 cells that had been untreated or treated with Y27632, a well known specific inhibitor for Rock kinase activity. We show here that the Rock pathway promotes a weak impact on the fibroblast transcriptome, since its inhibition only results in changes in the expression of 2.3% of all the genes surveyed in the microarrays. Most Y27632-dependent genes are downregulated at moderate levels, indicating that the Rock pathway predominantly induces the upregulation of transcriptionally active genes. Although functionally diverse, a common functional leitmotiv of Y27632-dependent genes is the implication of their protein products in cytoskeletal-dependent processes. Taken together, these results indicate that Rock proteins can modify cytoskeletal dynamics by acting at post-transcriptional and transcriptional levels. In addition, they suggest that the main target of these serine/threonine kinases is the phosphoproteome and not the transcriptome.
Figures


References
-
- Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. - PubMed
-
- Symons M, Rusk N. Control of vesicular trafficking by Rho GTPases. Curr Biol. 2003;13:R409–R418. - PubMed
-
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. - PubMed
-
- Coleman ML, Marshall CJ, Olson MF. RAS and RHO GTPases in G1-phase cell-cycle regulation. Nat Rev Mol Cell Biol. 2004;5:355–366. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
