Characterization of a highly conserved binding site of Mlh1 required for exonuclease I-dependent mismatch repair
- PMID: 19015241
- PMCID: PMC2630692
- DOI: 10.1128/MCB.00945-08
Characterization of a highly conserved binding site of Mlh1 required for exonuclease I-dependent mismatch repair
Abstract
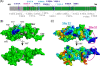
Mlh1 is an essential factor of mismatch repair (MMR) and meiotic recombination. It interacts through its C-terminal region with MutL homologs and proteins involved in DNA repair and replication. In this study, we identified the site of yeast Mlh1 critical for the interaction with Exo1, Ntg2, and Sgs1 proteins, designated as site S2 by reference to the Mlh1/Pms1 heterodimerization site S1. We show that site S2 is also involved in the interaction between human MLH1 and EXO1 or BLM. Binding at this site involves a common motif on Mlh1 partners that we called the MIP-box for the Mlh1 interacting protein box. Direct and specific interactions between yeast Mlh1 and peptides derived from Exo1, Ntg2, and Sgs1 and between human MLH1 and peptide derived from EXO1 and BLM were measured with K(d) values ranging from 8.1 to 17.4 microM. In Saccharomyces cerevisiae, a mutant of Mlh1 targeted at site S2 (Mlh1-E682A) behaves as a hypomorphic form of Exo1. The site S2 in Mlh1 mediates Exo1 recruitment in order to optimize MMR-dependent mutation avoidance. Given the conservation of Mlh1 and Exo1 interaction, it may readily impact Mlh1-dependent functions such as cancer prevention in higher eukaryotes.
Figures

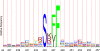



References
-
- Baker, S. M., A. W. Plug, T. A. Prolla, C. E. Bronner, A. C. Harris, X. Yao, D. M. Christie, C. Monell, N. Arnheim, A. Bradley, T. Ashley, and R. M. Liskay. 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13336-342. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
