Localization of the domains of the Haemophilus ducreyi trimeric autotransporter DsrA involved in serum resistance and binding to the extracellular matrix proteins fibronectin and vitronectin
- PMID: 19015257
- PMCID: PMC2632028
- DOI: 10.1128/IAI.00819-08
Localization of the domains of the Haemophilus ducreyi trimeric autotransporter DsrA involved in serum resistance and binding to the extracellular matrix proteins fibronectin and vitronectin
Abstract
Resisting the bactericidal activity of naturally occurring antibodies and complement of normal human serum is an important element in the evasion of innate immunity by bacteria. In the gram-negative mucosal pathogen Haemophilus ducreyi, serum resistance is mediated primarily by the trimeric autotransporter DsrA. DsrA also functions as an adhesin for the extracellular matrix proteins fibronectin and vitronectin and mediates attachment of H. ducreyi to keratinocytes. We sought to determine the domain(s) of the 236-residue DsrA protein required for serum resistance and extracellular matrix protein binding. A 140-amino-acid truncated protein containing only the C-terminal portion of the passenger domain and the entire translocator domain of DsrA exhibited binding to fibronectin and vitronectin and conferred serum resistance to an H. ducreyi serum-sensitive strain. A shorter DsrA construct consisting of only 128 amino acids was unable to bind to extracellular matrix proteins but was serum resistant. We concluded that neither fibronectin binding nor vitronectin binding is required for high-level serum resistance in H. ducreyi.
Figures


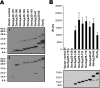

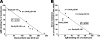
Similar articles
-
Defining Potential Vaccine Targets of Haemophilus ducreyi Trimeric Autotransporter Adhesin DsrA.Monoclon Antib Immunodiagn Immunother. 2015 Apr;34(2):73-82. doi: 10.1089/mab.2014.0054. Monoclon Antib Immunodiagn Immunother. 2015. PMID: 25897604 Free PMC article.
-
Outer membrane protein DsrA is the major fibronectin-binding determinant of Haemophilus ducreyi.Infect Immun. 2008 Apr;76(4):1608-16. doi: 10.1128/IAI.00994-07. Epub 2008 Jan 22. Infect Immun. 2008. PMID: 18212073 Free PMC article.
-
Trimeric autotransporter DsrA is a major mediator of fibrinogen binding in Haemophilus ducreyi.Infect Immun. 2013 Dec;81(12):4443-52. doi: 10.1128/IAI.00743-13. Epub 2013 Sep 16. Infect Immun. 2013. PMID: 24042118 Free PMC article.
-
The Haemophilus ducreyi serum resistance antigen DsrA confers attachment to human keratinocytes.Infect Immun. 2002 Nov;70(11):6158-65. doi: 10.1128/IAI.70.11.6158-6165.2002. Infect Immun. 2002. PMID: 12379693 Free PMC article.
-
The molecular basis of fibronectin-mediated bacterial adherence to host cells.Mol Microbiol. 2004 May;52(3):631-41. doi: 10.1111/j.1365-2958.2004.04027.x. Mol Microbiol. 2004. PMID: 15101971 Review.
Cited by
-
Attachment of Actinobacillus suis H91-0380 and Its Isogenic Adhesin Mutants to Extracellular Matrix Components of the Tonsils of the Soft Palate of Swine.Infect Immun. 2016 Sep 19;84(10):2944-52. doi: 10.1128/IAI.00456-16. Print 2016 Oct. Infect Immun. 2016. PMID: 27481253 Free PMC article.
-
An Acinetobacter trimeric autotransporter adhesin reaped from cells exhibits its nonspecific stickiness via a highly stable 3D structure.Sci Rep. 2016 Jun 16;6:28020. doi: 10.1038/srep28020. Sci Rep. 2016. PMID: 27305955 Free PMC article.
-
Virulence-associated trimeric autotransporters of Haemophilus parasuis are antigenic proteins expressed in vivo.Vet Res. 2010 May-Jun;41(3):26. doi: 10.1051/vetres/2009074. Epub 2009 Dec 10. Vet Res. 2010. PMID: 19995512 Free PMC article.
-
BBK32 attenuates antibody-dependent complement-mediated killing of infectious Borreliella burgdorferi isolates.PLoS Pathog. 2025 Jul 24;21(7):e1013361. doi: 10.1371/journal.ppat.1013361. eCollection 2025 Jul. PLoS Pathog. 2025. PMID: 40705830 Free PMC article.
-
Immunization with the Haemophilus ducreyi trimeric autotransporter adhesin DsrA with alum, CpG or imiquimod generates a persistent humoral immune response that recognizes the bacterial surface.Vaccine. 2016 Feb 24;34(9):1193-200. doi: 10.1016/j.vaccine.2016.01.024. Epub 2016 Jan 24. Vaccine. 2016. PMID: 26812077 Free PMC article.
References
-
- Ackermann, N., M. Tiller, G. Anding, A. Roggenkamp, and J. Heesemann. 2008. Contribution of trimeric autotransporter C-terminal domains of oligomeric coiled-coil adhesin (Oca) family members YadA, UspA1, EibA, and Hia to translocation of the YadA passenger domain and virulence of Yersinia enterocolitica. J. Bacteriol. 1905031-5043. - PMC - PubMed
-
- Al-Tawfiq, J. A., A. C. Thornton, B. P. Katz, K. R. Fortney, K. D. Todd, A. F. Hood, and S. M. Spinola. 1998. Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J. Infect. Dis. 1781684-1687. - PubMed
-
- Reference deleted.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

