Structural and biochemical studies of TIGAR (TP53-induced glycolysis and apoptosis regulator)
- PMID: 19015259
- PMCID: PMC2615519
- DOI: 10.1074/jbc.M807821200
Structural and biochemical studies of TIGAR (TP53-induced glycolysis and apoptosis regulator)
Abstract
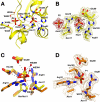
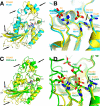
Activation of the p53 tumor suppressor by cellular stress leads to variable responses ranging from growth inhibition to apoptosis. TIGAR is a novel p53-inducible gene that inhibits glycolysis by reducing cellular levels of fructose-2,6-bisphosphate, an activator of glycolysis and inhibitor of gluconeogenesis. Here we describe structural and biochemical studies of TIGAR from Danio rerio. The overall structure forms a histidine phosphatase fold with a phosphate molecule coordinated to the catalytic histidine residue and a second phosphate molecule in a position not observed in other phosphatases. The recombinant human and zebra fish enzymes hydrolyze fructose-2,6-bisphosphate as well as fructose-1,6-bisphosphate but not fructose 6-phosphate in vitro. The TIGAR active site is open and positively charged, consistent with its enzymatic function as bisphosphatase. The closest related structures are the bacterial broad specificity phosphatase PhoE and the fructose-2,6-bisphosphatase domain of the bifunctional 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. The structural comparison shows that TIGAR combines an accessible active site as observed in PhoE with a charged substrate-binding pocket as seen in the fructose-2,6-bisphosphatase domain of the bifunctional enzyme.
Figures

References
-
- Laptenko, O., and Prives, C. (2006) Cell Death Differ. 13 951-961 - PubMed
-
- Vogelstein, B., and Kinzler, K. W. (2004) Nat. Med. 10 789-799 - PubMed
-
- Harris, S. L., and Levine, A. J. (2005) Oncogene 24 2899-2908 - PubMed
-
- Bensaad, K., Tsuruta, A., Selak, M. A., Vidal, M. N. C., Nakano, K., Bartrons, R., Gottlieb, E., and Vousden, K. H. (2006) Cell 126 107-120 - PubMed
-
- Michels, P. A. M., and Rigden, D. J. (2006) IUBMB Life 58 133-141 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

