Molecular structural basis for polymorphism in Alzheimer's beta-amyloid fibrils
- PMID: 19015532
- PMCID: PMC2587602
- DOI: 10.1073/pnas.0806270105
Molecular structural basis for polymorphism in Alzheimer's beta-amyloid fibrils
Abstract
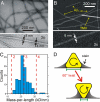
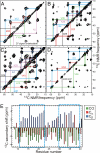
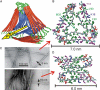
We describe a full structural model for amyloid fibrils formed by the 40-residue beta-amyloid peptide associated with Alzheimer's disease (Abeta(1-40)), based on numerous constraints from solid state NMR and electron microscopy. This model applies specifically to fibrils with a periodically twisted morphology, with twist period equal to 120 +/- 20 nm (defined as the distance between apparent minima in fibril width in negatively stained transmission electron microscope images). The structure has threefold symmetry about the fibril growth axis, implied by mass-per-length data and the observation of a single set of (13)C NMR signals. Comparison with a previously reported model for Abeta(1-40) fibrils with a qualitatively different, striated ribbon morphology reveals the molecular basis for polymorphism. At the molecular level, the 2 Abeta(1-40) fibril morphologies differ in overall symmetry (twofold vs. threefold), the conformation of non-beta-strand segments, and certain quaternary contacts. Both morphologies contain in-register parallel beta-sheets, constructed from nearly the same beta-strand segments. Because twisted and striated ribbon morphologies are also observed for amyloid fibrils formed by other polypeptides, such as the amylin peptide associated with type 2 diabetes, these structural variations may have general implications.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid: From bacteria to humans. Trends BiochemSci. 2007;32:217–224. - PubMed
-
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. - PubMed
-
- Tycko R. Molecular structure of amyloid fibrils: Insights from solid state NMR. Q Rev Biophys. 2006;39:1–55. - PubMed
-
- Petkova AT, et al. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 2005;307:262–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

