Probing protein structure by amino acid-specific covalent labeling and mass spectrometry
- PMID: 19016300
- PMCID: PMC2768138
- DOI: 10.1002/mas.20203
Probing protein structure by amino acid-specific covalent labeling and mass spectrometry
Abstract
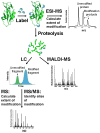
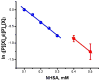
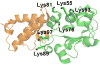
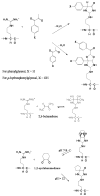
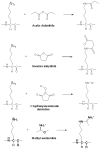
For many years, amino acid-specific covalent labeling has been a valuable tool to study protein structure and protein interactions, especially for systems that are difficult to study by other means. These covalent labeling methods typically map protein structure and interactions by measuring the differential reactivity of amino acid side chains. The reactivity of amino acids in proteins generally depends on the accessibility of the side chain to the reagent, the inherent reactivity of the label and the reactivity of the amino acid side chain. Peptide mass mapping with ESI- or MALDI-MS and peptide sequencing with tandem MS are typically employed to identify modification sites to provide site-specific structural information. In this review, we describe the reagents that are most commonly used in these residue-specific modification reactions, details about the proper use of these covalent labeling reagents, and information about the specific biochemical problems that have been addressed with covalent labeling strategies.
Copyright 2008 Wiley Periodicals, Inc.
Figures



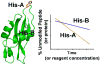
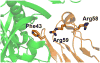



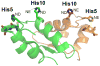







References
-
- Akashi S, Shirouzu M, Terada T, Ito Y, Yokohama S, Takio K. Characterization of the structural difference between active and inactive forms of the Ras protein by chemical modification followed by mass spectrometric peptide mapping. Anal Biochem. 1997;248:15–25. - PubMed
-
- Akinsiku OT, Yu ET, Fabris D. Mass spectrometric investigation of protein alkylation by the RNA footprinting probe kethoxal. J Mass Spectrom. 2005;40:1372–1381. - PubMed
-
- Apuy JL, Chen XH, Russell DH, Baldwin TO, Giedroc DP. Ratiometric pulsed Alkylation/Mass spectrometry of the cysteine pairs in individual zinc fingers of MRE-Binding transcription factor-1 (MTF-1) as a probe of zinc chelate stability. Biochemistry. 2001;40:15164–15175. - PubMed
-
- Apuy JL, Park ZY, Swartz PD, Dangott LJ, Russell DH, Baldwin TO. Pulsed-alkylation mass spectrometry for the study of protein folding and dynamics: Development and application to the study of a folding/unfolding intermediate of bacterial luciferaset. Biochemistry. 2001;40:15153–15163. - PubMed
-
- Azim-Zadeh O, Hillebrecht A, Linne U, Marahiel MA, Klebe G, Lingelbach K, Nyalwidhe J. Use of biotin derivatives to probe conformational changes in proteins. J Biol Chem. 2007;282:21609–21617. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

