Bacteria as computers making computers
- PMID: 19016882
- PMCID: PMC2704931
- DOI: 10.1111/j.1574-6976.2008.00137.x
Bacteria as computers making computers
Abstract
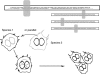
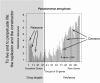
Various efforts to integrate biological knowledge into networks of interactions have produced a lively microbial systems biology. Putting molecular biology and computer sciences in perspective, we review another trend in systems biology, in which recursivity and information replace the usual concepts of differential equations, feedback and feedforward loops and the like. Noting that the processes of gene expression separate the genome from the cell machinery, we analyse the role of the separation between machine and program in computers. However, computers do not make computers. For cells to make cells requires a specific organization of the genetic program, which we investigate using available knowledge. Microbial genomes are organized into a paleome (the name emphasizes the role of the corresponding functions from the time of the origin of life), comprising a constructor and a replicator, and a cenome (emphasizing community-relevant genes), made up of genes that permit life in a particular context. The cell duplication process supposes rejuvenation of the machine and replication of the program. The paleome also possesses genes that enable information to accumulate in a ratchet-like process down the generations. The systems biology must include the dynamics of information creation in its future developments.
Figures

References
-
- Allen C, Bekoff M, Lauder G. Nature's Purposes. Cambridge, MA: MIT Press; 1998.
-
- Alon U. An Introduction to Systems Biology. Design Principles of Biological Circuits. London: Chapman & Hall; 2006.
-
- Ara K, Ozaki K, Nakamura K, Yamane K, Sekiguchi J, Ogasawara N. Bacillus minimum genome factory: effective utilization of microbial genome information. Biotechnol Appl Bioc. 2007;46:169–178. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

