Multiple PLDs required for high salinity and water deficit tolerance in plants
- PMID: 19017627
- PMCID: PMC2638713
- DOI: 10.1093/pcp/pcn173
Multiple PLDs required for high salinity and water deficit tolerance in plants
Abstract
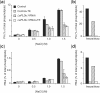
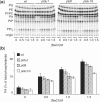
High salinity and drought have received much attention because they severely affect crop production worldwide. Analysis and comprehension of the plant's response to excessive salt and dehydration will aid in the development of stress-tolerant crop varieties. Signal transduction lies at the basis of the response to these stresses, and numerous signaling pathways have been implicated. Here, we provide further evidence for the involvement of phospholipase D (PLD) in the plant's response to high salinity and dehydration. A tomato (Lycopersicon esculentum) alpha-class PLD, LePLDalpha1, is transcriptionally up-regulated and activated in cell suspension cultures treated with salt. Gene silencing revealed that this PLD is indeed involved in the salt-induced phosphatidic acid production, but not exclusively. Genetically modified tomato plants with reduced LePLDalpha1 protein levels did not reveal altered salt tolerance. In Arabidopsis (Arabidopsis thaliana), both AtPLDalpha1 and AtPLDdelta were found to be activated in response to salt stress. Moreover, pldalpha1 and plddelta single and double knock-out mutants exhibited enhanced sensitivity to high salinity stress in a plate assay. Furthermore, we show that both PLDs are activated upon dehydration and the knock-out mutants are hypersensitive to hyperosmotic stress, displaying strongly reduced growth.
Figures






References
-
- Bargmann BOR, Munnik T. The role of phospholipase D in plant stress responses. Curr. Opin. Plant Biol. 2006;9:515–522. - PubMed
-
- Bargmann BOR, Laxalt AM, ter Riet B, Schouten E, van Leeuwen W, Dekker HL, et al. LePLDbeta1 activation and relocalization in suspension-cultured tomato cells treated with xylanase. Plant J. 2006;45:58–68. - PubMed
-
- Boudsocq M, Barbier-Brygoo H, Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004;279:41758–41766. - PubMed
-
- Burza AM, Pekala I, Sikora J, Siedlecki P, Małagocki P, Bucholc M, et al. Nicotiana tabacum osmotic stress-activated kinase is regulated by phosphorylation on Ser-154 and Ser-158 in the kinase activation loop. J. Biol. Chem. 2006;281:34299–34311. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

