Differential nuclear scaffold/matrix attachment marks expressed genes
- PMID: 19017725
- PMCID: PMC2638830
- DOI: 10.1093/hmg/ddn394
Differential nuclear scaffold/matrix attachment marks expressed genes
Abstract
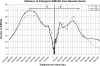
It is well established that nuclear architecture plays a key role in poising regions of the genome for transcription. This may be achieved using scaffold/matrix attachment regions (S/MARs) that establish loop domains. However, the relationship between changes in the physical structure of the genome as mediated by attachment to the nuclear scaffold/matrix and gene expression is not clearly understood. To define the role of S/MARs in organizing our genome and to resolve the often contradictory loci-specific studies, we have surveyed the S/MARs in HeLa S3 cells on human chromosomes 14-18 by array comparative genomic hybridization. Comparison of LIS (lithium 3,5-diiodosalicylate) extraction to identify SARs and 2 m NaCl extraction to identify MARs revealed that approximately one-half of the sites were in common. The results presented in this study suggest that SARs 5' of a gene are associated with transcript presence whereas MARs contained within a gene are associated with silenced genes. The varied functions of the S/MARs as revealed by the different extraction methods highlights their unique functional contribution.
Figures
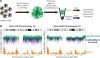



References
-
- Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. - PubMed
-
- Volpi E.V., Chevret E., Jones T., Vatcheva R., Williamson J., Beck S., Campbell R.D., Goldsworthy M., Powis S.H., Ragoussis J., et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 2000;113(Pt):1565–1576. - PubMed
-
- Albiez H., Cremer M., Tiberi C., Vecchio L., Schermelleh L., Dittrich S., Kupper K., Joffe B., Thormeyer T., von Hase J., et al. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 2006;14:707–733. - PubMed
-
- Bode J., Benham C., Knopp A., Mielke C. Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements) Crit. Rev. Eukaryot. Gene Expr. 2000;10:73–90. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

