The replisome uses mRNA as a primer after colliding with RNA polymerase
- PMID: 19020502
- PMCID: PMC2605185
- DOI: 10.1038/nature07527
The replisome uses mRNA as a primer after colliding with RNA polymerase
Abstract
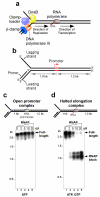
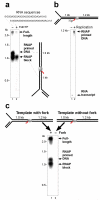
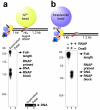
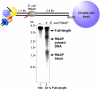
Replication forks are impeded by DNA damage and protein-nucleic acid complexes such as transcribing RNA polymerase. For example, head-on collision of the replisome with RNA polymerase results in replication fork arrest. However, co-directional collision of the replisome with RNA polymerase has little or no effect on fork progression. Here we examine co-directional collisions between a replisome and RNA polymerase in vitro. We show that the Escherichia coli replisome uses the RNA transcript as a primer to continue leading-strand synthesis after the collision with RNA polymerase that is displaced from the DNA. This action results in a discontinuity in the leading strand, yet the replisome remains intact and bound to DNA during the entire process. These findings underscore the notable plasticity by which the replisome operates to circumvent obstacles in its path and may explain why the leading strand is synthesized discontinuously in vivo.
Figures

References
-
- Rudolph CJ, Dhillon P, Moore T, Lloyd RG. Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair (Amst) 2007;6(7):981–993. - PubMed
-
- Cox MM, et al. The importance of repairing stalled replication forks. Nature. 2000;404(6773):37–41. - PubMed
-
- Cox MM. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu Rev Genet. 2001;35:53–82. - PubMed
-
- Brewer BJ. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53(5):679–686. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

