Iron-based redox switches in biology
- PMID: 19021503
- PMCID: PMC2842161
- DOI: 10.1089/ars.2008.2296
Iron-based redox switches in biology
Abstract
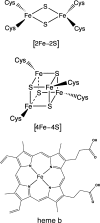
By virtue of its unique electrochemical properties, iron makes an ideal redox active cofactor for many biologic processes. In addition to its important role in respiration, central metabolism, nitrogen fixation, and photosynthesis, iron also is used as a sensor of cellular redox status. Iron-based sensors incorporate Fe-S clusters, heme, and mononuclear iron sites to act as switches to control protein activity in response to changes in cellular redox balance. Here we provide an overview of iron-based redox sensor proteins, in both prokaryotes and eukaryotes, that have been characterized at the biochemical level. Although this review emphasizes redox sensors containing Fe-S clusters, proteins that use heme or novel iron sites also are discussed.
Figures



References
-
- Althaus EW. Outten CE. Olson KE. Cao H. O'Halloran TV. The ferric uptake regulation (Fur) repressor is a zinc metalloprotein. Biochemistry. 1999;38:6559–6569. - PubMed
-
- Andrews SC. Robinson AK. Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. - PubMed
-
- Anxolabehere-Mallart E. Glaser T. Frank P. Aliverti A. Zanetti G. Hedman B. Hodgson KO. Solomon EI. Sulfur K-edge X-ray absorption spectroscopy of 2Fe-2S ferredoxin: covalency of the oxidized and reduced 2Fe forms and comparison to model complexes. J Am Chem Soc. 2001;123:5444–5452. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

