Exploring the mialome of ticks: an annotated catalogue of midgut transcripts from the hard tick, Dermacentor variabilis (Acari: Ixodidae)
- PMID: 19021911
- PMCID: PMC2644717
- DOI: 10.1186/1471-2164-9-552
Exploring the mialome of ticks: an annotated catalogue of midgut transcripts from the hard tick, Dermacentor variabilis (Acari: Ixodidae)
Abstract
Background: Ticks are obligate blood feeders. The midgut is the first major region of the body where blood and microbes ingested with the blood meal come in contact with the tick's internal tissues. Little is known about protein expression in the digestive tract of ticks. In this study, for analysis of global gene expression during tick attachment and feeding, we generated and sequenced 1,679 random transcripts (ESTs) from cDNA libraries from the midguts of female ticks at varying stages of feeding.
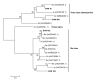
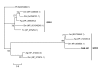
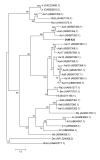
Results: Sequence analysis of the 1,679 ESTs resulted in the identification of 835 distinct transcripts, from these, a total of 82 transcripts were identified as proteins putatively directly involved in blood meal digestion, including enzymes involved in oxidative stress reduction/antimicrobial activity/detoxification, peptidase inhibitors, protein digestion (cysteine-, aspartic-, serine-, and metallo-peptidases), cell, protein and lipid binding including mucins and iron/heme metabolism and transport. A lectin-like protein with a high match to lectins in other tick species, allergen-like proteins and surface antigens important in pathogen recognition and/or antimicrobial activity were also found. Furthermore, midguts collected from the 6-day-fed ticks expressed twice as many transcripts involved in bloodmeal processing as midguts from unfed/2-day-fed ticks.
Conclusion: This tissue-specific transcriptome analysis provides an opportunity to examine the global expression of transcripts in the tick midgut and to compare the gut response to host attachment versus blood feeding and digestion. In contrast to those in salivary glands of other Ixodid ticks, most proteins in the D. variabilis midgut cDNA library were intracellular. Of the total ESTs associated with a function, an unusually large number of transcripts were associated with peptidases, cell, lipid and protein binding, and oxidative stress or detoxification. Presumably, this is consistent with their role in intracellular processing of the blood meal and response to microbial infections. The presence of many proteins with similar functions is consistent with the hypothesis that gene duplication contributed to the successful adaptation of ticks to hematophagy. Furthermore, these transcripts may be useful to scientists investigating the role of the tick midgut in blood-meal digestion, antimicrobial activity or the transmission of tick-borne pathogens.
Figures
















References
-
- Fogaca AC, da Silva PI, Jr, Miranda MT, Bianchi AG, Miranda A, Ribolla PE, Daffre S. Antimicrobial activity of a bovine hemoglobin fragment in the tick Boophilus microplus. J Biol Chem. 1999;274:25330–25334. - PubMed
-
- Nakajima Y, Ogihara K, Taylor D, Yamakawa M. Antibacterial hemoglobin fragments from the midgut of the soft tick, Ornithodoros moubata (Acari: Argasidae) J Med Entomol. 2003;40:78–81. - PubMed
-
- Sonenshine DE, Hynes WL, Ceraul SM, Mitchell R, Benzine T. Host blood proteins and peptides in the midgut of the tick Dermacentor variabilis contribute to bacterial control. Exp Appl Acarol. 2005;36:207–223. - PubMed
-
- Nakajima Y, Goes van Naters-Yasui A van der, Taylor D, Yamakawa M. Antibacterial peptide defensin is involved in midgut immunity of the soft tick, Ornithodoros moubata. Insect Mol Biol. 2002;11:611–618. - PubMed
-
- Kopacek P, Vogt R, Jindrak L, Weise C, Safarik I. Purification and characterization of the lysozyme from the gut of the soft tick Ornithodoros moubata. Insect Biochem Mol Biol. 1999;29:989–997. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

