the hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin
- PMID: 19023418
- PMCID: PMC2581891
- DOI: 10.1371/journal.ppat.1000217
the hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin
Abstract
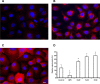
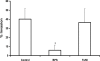
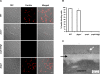
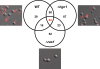
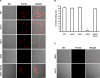
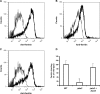
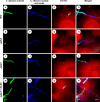
Iron sequestration by host iron-binding proteins is an important mechanism of resistance to microbial infections. Inside oral epithelial cells, iron is stored within ferritin, and is therefore not usually accessible to pathogenic microbes. We observed that the ferritin concentration within oral epithelial cells was directly related to their susceptibility to damage by the human pathogenic fungus, Candida albicans. Thus, we hypothesized that host ferritin is used as an iron source by this organism. We found that C. albicans was able to grow on agar at physiological pH with ferritin as the sole source of iron, while the baker's yeast Saccharomyces cerevisiae could not. A screen of C. albicans mutants lacking components of each of the three known iron acquisition systems revealed that only the reductive pathway is involved in iron utilization from ferritin by this fungus. Additionally, C. albicans hyphae, but not yeast cells, bound ferritin, and this binding was crucial for iron acquisition from ferritin. Transcriptional profiling of wild-type and hyphal-defective C. albicans strains suggested that the C. albicans invasin-like protein Als3 is required for ferritin binding. Hyphae of an Deltaals3 null mutant had a strongly reduced ability to bind ferritin and these mutant cells grew poorly on agar plates with ferritin as the sole source of iron. Heterologous expression of Als3, but not Als1 or Als5, two closely related members of the Als protein family, allowed S. cerevisiae to bind ferritin. Immunocytochemical localization of ferritin in epithelial cells infected with C. albicans showed ferritin surrounding invading hyphae of the wild-type, but not the Deltaals3 mutant strain. This mutant was also unable to damage epithelial cells in vitro. Therefore, C. albicans can exploit iron from ferritin via morphology dependent binding through Als3, suggesting that this single protein has multiple virulence attributes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. - PubMed
-
- Howard DH. Iron gathering by zoopathogenic fungi. FEMS Immunol Med Microbiol. 2004;40:95–100. - PubMed
-
- Ekins A, Khan AG, Shouldice SR, Schryvers AB. Lactoferrin receptors in gram-negative bacteria: insights into the iron acquisition process. Biometals. 2004;17:235–243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

