Genomic islands of Pseudomonas aeruginosa
- PMID: 19025565
- PMCID: PMC2648531
- DOI: 10.1111/j.1574-6968.2008.01406.x
Genomic islands of Pseudomonas aeruginosa
Abstract
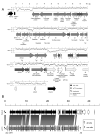
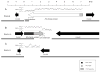
Key to Pseudomonas aeruginosa's ability to thrive in a diversity of niches is the presence of numerous genomic islands that confer adaptive traits upon individual strains. We reasoned that P. aeruginosa strains capable of surviving in the harsh environments of multiple hosts would therefore represent rich sources of genomic islands. To this end, we identified a strain, PSE9, that was virulent in both animals and plants. Subtractive hybridization was used to compare the genome of PSE9 with the less virulent strain PAO1. Nine genomic islands were identified in PSE9 that were absent in PAO1; seven of these had not been described previously. One of these seven islands, designated P. aeruginosa genomic island (PAGI)-5, has already been shown to carry numerous interesting ORFs, including several required for virulence in mammals. Here we describe the remaining six genomic islands, PAGI-6, -7, -8, -9, -10, and -11, which include a prophage element and two Rhs elements.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Cheetham BF, Katz ME. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18:201–208. - PubMed
-
- Costas AM, White AK, Metcalf WW. Purification and characterization of a novel phosphorus-oxidizing enzyme from Pseudomonas stutzeri WM88. J Biol Chem. 2001;276:17429–17436. - PubMed
-
- Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

