All-paths graph kernel for protein-protein interaction extraction with evaluation of cross-corpus learning
- PMID: 19025688
- PMCID: PMC2586751
- DOI: 10.1186/1471-2105-9-S11-S2
All-paths graph kernel for protein-protein interaction extraction with evaluation of cross-corpus learning
Abstract
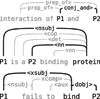
Background: Automated extraction of protein-protein interactions (PPI) is an important and widely studied task in biomedical text mining. We propose a graph kernel based approach for this task. In contrast to earlier approaches to PPI extraction, the introduced all-paths graph kernel has the capability to make use of full, general dependency graphs representing the sentence structure.
Results: We evaluate the proposed method on five publicly available PPI corpora, providing the most comprehensive evaluation done for a machine learning based PPI-extraction system. We additionally perform a detailed evaluation of the effects of training and testing on different resources, providing insight into the challenges involved in applying a system beyond the data it was trained on. Our method is shown to achieve state-of-the-art performance with respect to comparable evaluations, with 56.4 F-score and 84.8 AUC on the AImed corpus.
Conclusion: We show that the graph kernel approach performs on state-of-the-art level in PPI extraction, and note the possible extension to the task of extracting complex interactions. Cross-corpus results provide further insight into how the learning generalizes beyond individual corpora. Further, we identify several pitfalls that can make evaluations of PPI-extraction systems incomparable, or even invalid. These include incorrect cross-validation strategies and problems related to comparing F-score results achieved on different evaluation resources. Recommendations for avoiding these pitfalls are provided.
Figures

References
-
- Hirschman L, Park JC, Tsujii J, Wong L, Wu CH. Accomplishments and challenges in literature data mining for biology. Bioinformatics. 2002;18:1553–1561. - PubMed
-
- Cohen KB, Hunter L. Artificial intelligence methods and tools for systems biology, Volume 5 of Computational Biology. Springer; 2004. Natural language processing and systems biology; pp. 147–173.
-
- Bunescu R, Ge R, Kate R, Marcotte E, Mooney R, Ramani A, Wong Y. Comparative Experiments on Learning Information Extractors for Proteins and their Interactions. Artificial Intelligence in Medicine. 2005;33:139–155. - PubMed
-
- Hunter L, Lu Z, Firby J, Baumgartner WA, Johnson HL, Ogren PV, Cohen KB. OpenDMAP: An open-source, ontology-driven concept analysis engine, with applications to capturing knowledge regarding protein transport, protein interactions and cell-specific gene expression. BMC Bioinformatics. 2008;9:78. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

