Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein
- PMID: 19028679
- PMCID: PMC2631945
- DOI: 10.1074/jbc.M808032200
Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein
Abstract
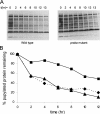
The striking identification of an apparent proteasome core in Mycobacteria and allied actinomycetes suggested that additional elements of this otherwise strictly eukaryotic system for regulated protein degradation might be conserved. The genes encoding this prokaryotic proteasome are clustered in an operon with a short open reading frame that encodes a small protein of 64 amino acids resembling ubiquitin with a carboxyl-terminal di-glycine-glutamine motif (herein called Pup for prokaryotic ubiquitin-like protein). Expression of a polyhistidine-tagged Pup followed by pulldown revealed that a broad spectrum of proteins were post-translationally modified by Pup. Two-dimensional gel electrophoresis allowed us to conclusively identify two targets of this modification as myoinositol-1-phosphate synthase and superoxide dismutase. Deletion of the penultimate di-glycine motif or the terminal glutamine completely abrogated modification of cellular proteins with Pup. Further mass spectral analysis demonstrated that Pup was attached to a lysine residue on its target protein via the carboxyl-terminal glutamine with deamidation of this residue. Finally, we showed that cell lysates of wild type (but not a proteasome mutant) efficiently degraded Pup-modified proteins. These data therefore establish that, despite differences in both sequence and target linkage, Pup plays an analogous role to ubiquitin in targeting proteins to the proteasome for degradation.
Figures



References
-
- Dahlmann, B., Kopp, F., Kuehn, L., Niedel, B., Pfeifer, G., Hegerl, R., and Baumeister, W. (1989) FEBS Lett. 251 125-131 - PubMed
-
- Lupas, A., Zwickl, P., and Baumeister, W. (1994) Trends Biochem. Sci. 19 533-534 - PubMed
-
- Tamura, T., Nagy, I., Lupas, A., Lottspeich, F., Cejka, Z., Schoofs, G., Tanaka, K., De Mot, R., and Baumeister, W. (1995) Curr. Biol. 5 766-774 - PubMed
-
- Cohen, R. E., and Pickart, C. M. (2004) Nat. Rev. Mol. Cell Biol. 5 177-187 - PubMed
-
- Glickman, M. H., Rubin, D. M., Coux, O., Wefes, I., Pfeifer, G., Cjeka, Z., Baumeister, W., Fried, V. A., and Finley, D. (1998) Cell 94 615-623 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

