EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates
- PMID: 19029536
- PMCID: PMC2652215
- DOI: 10.1101/gr.073585.107
EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates
Abstract
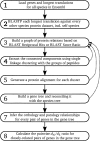
We have developed a comprehensive gene orientated phylogenetic resource, EnsemblCompara GeneTrees, based on a computational pipeline to handle clustering, multiple alignment, and tree generation, including the handling of large gene families. We developed two novel non-sequence-based metrics of gene tree correctness and benchmarked a number of tree methods. The TreeBeST method from TreeFam shows the best performance in our hands. We also compared this phylogenetic approach to clustering approaches for ortholog prediction, showing a large increase in coverage using the phylogenetic approach. All data are made available in a number of formats and will be kept up to date with the Ensembl project.
Figures




References
-
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., et al. The genome sequence of Drosophila melanogaster . Science. 2000;287:2185–2195. - PubMed
-
- Dehal P., Satou Y., Campbell R.K., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. - PubMed
-
- Dufayard J.F., Duret L., Penel S., Gouy M., Rechenmann F., Perriere G. Tree pattern matching in phylogenetic trees: Automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics. 2005;21:2596–2603. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
