Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae
- PMID: 19029899
- PMCID: PMC2596280
- DOI: 10.1038/ng.280
Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae
Abstract
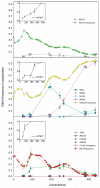
The classical model of adaptive evolution in an asexual population postulates that each adaptive clone is derived from the one preceding it. However, experimental evidence has suggested more complex dynamics, with theory predicting the fixation probability of a beneficial mutation as dependent on the mutation rate, population size and the mutation's selection coefficient. Clonal interference has been demonstrated in viruses and bacteria but not in a eukaryote, and a detailed molecular characterization is lacking. Here we use three different fluorescent markers to visualize the dynamics of asexually evolving yeast populations. For each adaptive clone within one of our evolving populations, we identified the underlying mutations, monitored their population frequencies and used microarrays to characterize changes in the transcriptome. These results represent the most detailed molecular characterization of experimental evolution to date and provide direct experimental evidence supporting both the clonal interference and the multiple mutation models.
Figures



References
-
- Muller HJ. Some genetic aspects of sex. American Naturalist. 1932;66:118–138.
-
- Notley-McRobb L, Ferenci T. The generation of multiple co-existing mal-regulatory mutations through polygenic evolution in glucose limited populations of Escherichia coli. Environmental Microbiology. 1999;1:45–52. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

